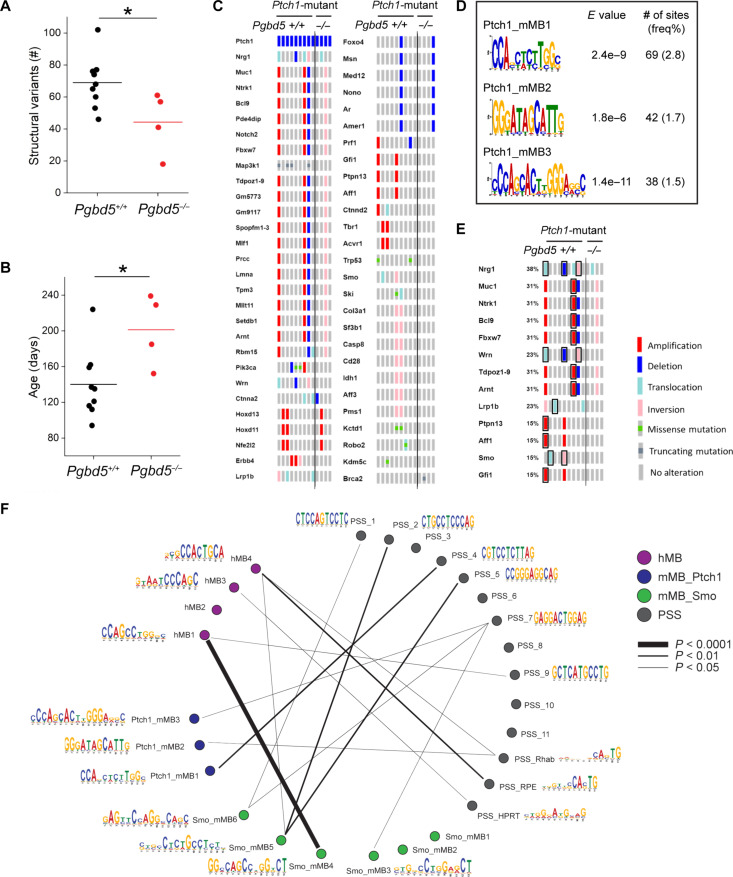
Fig. 3. Pgbd5 promotes somatic mutagenesis of recurrently mutated tumor suppressor and oncogenes in mouse SHH MBs.
(A) Numbers of SVs in Ptf1aCre/+;Ptch1fl/fl tumors. Pgbd5+/+ tumors (black, n = 9) harbor more SVs than Pgbd5−/− (red, n = 4). Lines indicate mean (69 and 44, respectively), and significance is measured using t test (*P = 0.036). (B) Age of tumors (days) in Ptch1-mutant tumors. Pgbd5+/+ tumors (black, n = 9) are younger than Pgbd5−/− tumors (red, n = 4; mean 140 and 201 days, *P = 0.024). (C) Oncoprint showing genes recurrently affected by SVs and SNVs in independent Ptch1-mutant tumors. Genes are curated based on likelihood that SVs or SNVs affect gene function (see Materials and Methods for details). The left nine and right four columns indicate tumors from Pgbd5+/+ and Pgbd5−/− mice, respectively. Red, blue, light blue, pink, and gray symbols indicate amplifications, deletions, translocations, inversions, and no alteration, respectively. Green and dark gray squares in gray symbols indicate missense and truncating mutations, respectively. (D) Three Pgbd5+/+-specific motifs are identified at SV breakpoints in Ptch1-mutant tumors, using discriminative MEME with Pgbd5−/− tumors as controls. E values indicate MEME discriminative algorithm significance (see Materials and Methods for details). The frequency shown was calculated by dividing the number of sites by total numbers of 50-bp breakpoint sequences extracted from SVs. (E) These motifs in (D) and the previously identified PSS_RPE and PSS_Rhab motifs were identified at SV breakpoints affecting known tumor suppressor and oncogenes in six of nine (66%) of Pgbd5+/+ tumors. The affected tumor suppressors and oncogenes are boxed black. (F) Circos plot showing similarities among all motifs (see Materials and Methods for details).