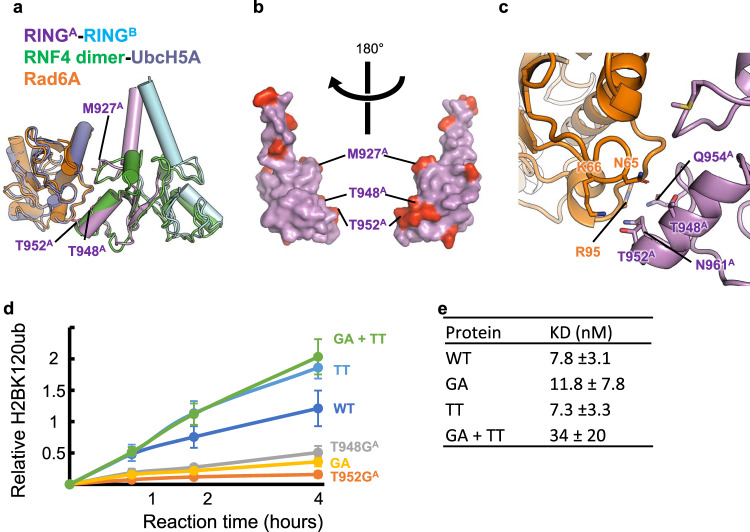
Fig. 4. Identification of Bre1A residues likely to mediate Rad6A binding.
a Structural modeling of the RINGA-RINGB-Rad6A complex. The current structure of the RINGA-RINGB complex is superposed on that of the RNF4 dimer bound to UbcH5A and ubiquitin (PDB ID 4AP4, ubiquitin not shown), and then the structure of Rad6A (PDB ID 6CYO) is further superposed on that of UbcH5A. b RINGA residues that are not conserved in RINGB. The RINGA structure is shown as a surface representation, and 10 non-conserved residues are colored red. Three residues near a possible Rad6A binding region are labeled. c Close-up view of the RINGA-RINGB-Rad6A model in (b). d H2BK120 ubiquitination assay of the wild type (WT) and mutants possessing substitutions at the possible Rad6A binding region. The constructs shown in Fig. 2d and their mutants were used. GA: T948GA-T952AA. TT: G974TB-A978TB. The mean and standard deviation of six (for WT) or three (for mutants) independent results are shown. Source data are provided as a Source Data file. e The binding affinities of the WT and mutants for the nucleosome. The constants between the Bre1 complexes and the nucleosome are shown with their 1-sigma confidence interval. The values were calculated with four (for GA + TT) or three (for the others) independent results.