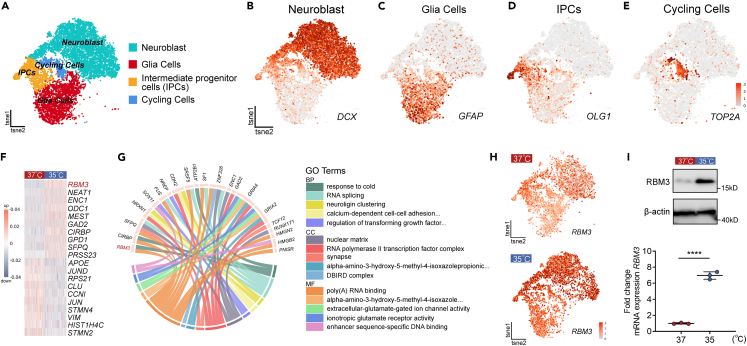
Figure 2.
RBM3 was identified as a potential regulator in neuronal differentiation of hNSCs by mild hypothermia
(A) The t-SNE plot of single-cell RNA sequencing showing individual cell types of differentiated hNSCs at Day 14 based on maker genes expression, including neuroblast, glia cells, intermediate progenitor cells and cycling cells.
(B–E) t-SNE plots highlighting cell types by marker genes expression: neuroblast (DCX), glia cells (GFAP), intermediate progenitor cells (OLIG1), and cycling cells (TOP2A).
(F) Heatmap of top 10 and last 10 differentially expressed genes (DEGs) between 35°C and 37°C in neuroblast clusters. Each row represents a gene, and each column represents a cell.
(G) Circos plot showing significant upregulated genes and GO analysis. The bottom half of circos plot shows the top 5 GO terms including BP, MF, CC and the top half shows the DEGs of the depicted GO enrichment analysis.
(H) t-SNE plots highlighting the RBM3 expression in differentiated hNSCs at Day 14 with 35°C- and 37°C-treatments. (I) Western blots and qRT-PCR analysis showing the protein and mRNA expressions of RBM3 in 35°C- and 37°C-treatments. β-actin was used as the loading control. All data presented as mean ± SD. Student’s t test was used in (I). ∗∗∗∗p < 0.0001.