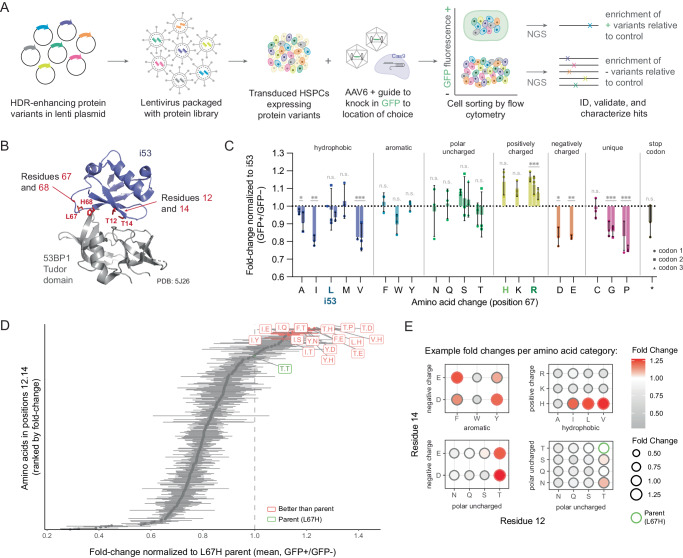
Fig. 1. A lentiviral-based pooled screening platform in HSPCs identifies HDR-enhancing variants of i53.
A Schematic outlining the lentiviral-based pooled screening platform in HSPCs used to identify protein-based additives to increase HDR at a Cas9-mediated cut site of interest using an AAV6 DNA donor template. Protein variants are encoded in lentiviral libraries; once integrated into the genome the sequences can be amplified from HSPC subpopulations. To functionally quantify homology-based repair in pooled libraries, transduced cells are edited using Cas9 RNP and an AAV6 template that encodes for a GFP insertion at the cut site of interest (e.g. HBB gene). Post editing (3–5 days), cells are sorted via flow cytometry into GFP+ and GFP- populations. Genomic DNA is extracted from each sorted cell population, sequenced via NGS, and analyzed to determine the distribution of variants relative to a control. B Residues targeted for mutagenesis were chosen by their proximity to the binding interface between with 53BP1 and i53 and are shown in red (T12, T14, L67, H68). C Example enrichment of residues following screening with a saturation mutagenesis (NNK) library at position L67 (parent: i53, library size = 32 unique codons encoding 20 variants). n = 3 separate pooled analyses and mean ± SD depicted. Each bar represents a unique codon for that amino acid. n.s. not significant; *p < 0.05; **p < 0.01; ***p < 0.001. Two-tailed t-test with Holm-Šídák correction for multiple comparisons. Exact p-values are reported on Source Data file. Of the 19 new variants tested, one (L67R) was found to be significantly enriched relative to parent i53 (L67), although L67H was borderline significant and was also moved on to subsequent validation. D Example enrichment of residues following a combinatorial library at positions T12 and T14 (parent: L67H), library size = 324 variants for which all replicates were enriched over parent are highlighted in red (16 variants, or 5%). n = 3 separate pooled analyses; bars represent mean ± SEM. Selected top hits were subsequently validated in focused libraries and experiments with purified recombinant protein. E Dot plot representation of variant fold change enrichment in combinatorial analysis, clustered by amino acid properties. Variations of residues 12 and 14 shown on the x-axis and y-axis, respectively. Additional information for this library shown in Supplementary Fig. 1.4. C–E Source data are provided as a Source Data file.