Figure 1 |. Research strategies for understanding the molecular basis of cancer metastasis: from reductionism to systems biology.
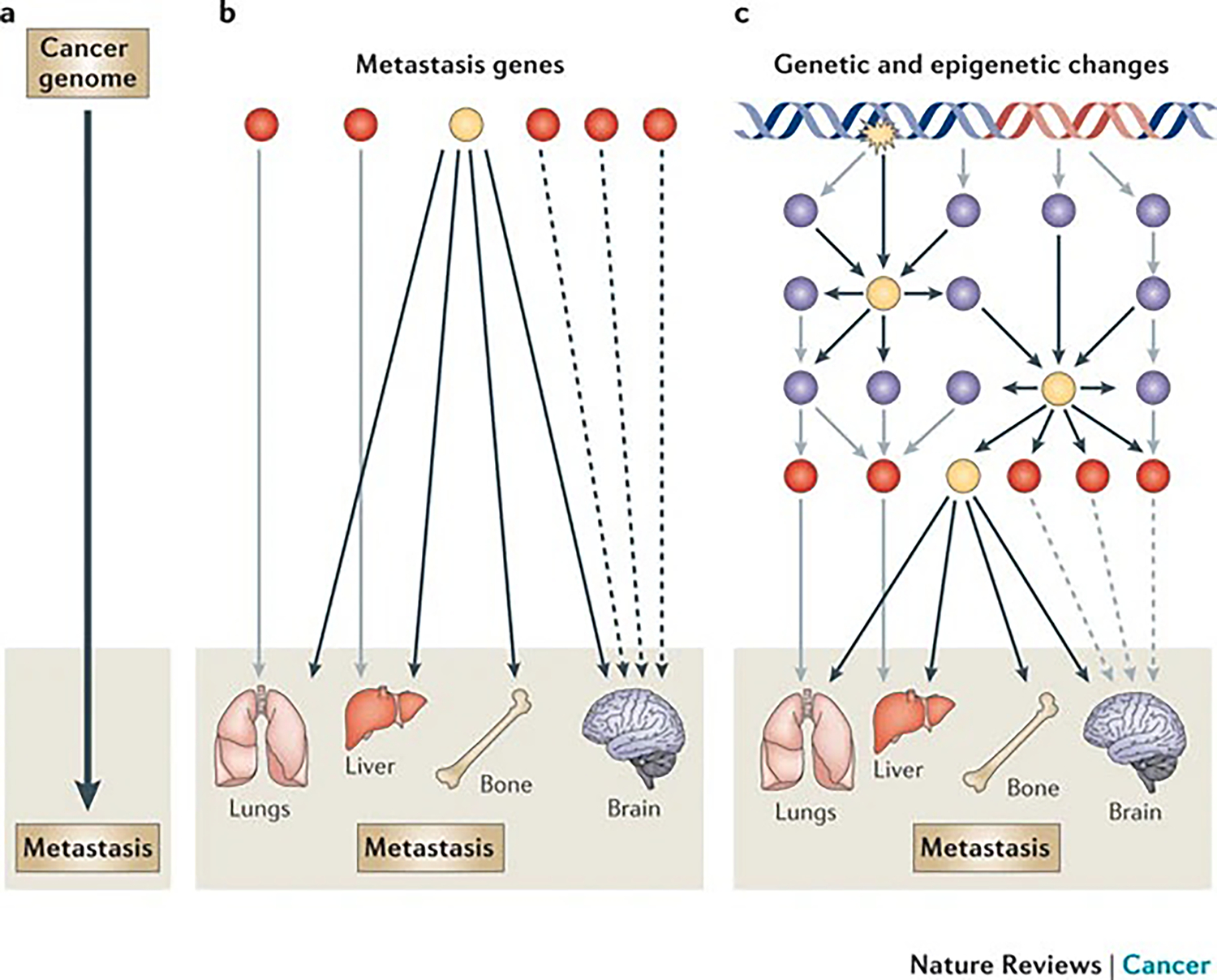
The evolving states of cancer, including metastasis, are reflected in dynamically changing expression patterns of the genes and proteins within the cancer cells. Metastasis research has generally relied on the linear approach of gene-to-trait mapping, which links the metastasis genotype with its corresponding metastatic phenotype (part a). More complex linear linkages may include multivalent relationships, in which one gene (or group of genes) can have functions in multiple metastasis phenotypes (pleiotropy), and a metastasis tissue tropism can be exerted by many independent genes or gene groups (redundancy) (part b). However, gene interactions are influenced by their context, which often cannot be captured by traditional one-gene-one-trait approaches. Therefore, metastatic behaviour should be considered as the consequence of the collective action of individual metastasis genes through nonlinear interactions (part c). Understanding the nature of these network level interactions and identifying crucial nodes of functional control will pave the way towards rational therapeutic design for metastatic breast cancer. Red circles represent genes or groups of genes that mediate tissue-specific metastasis of breast cancer. Blue circles represent regulators of metastasis genes. Yellow circles represent key functional nodes of metastasis regulation networks and are prime targets for therapeutic development. The black, grey and dashed arrows indicate different pathways.
