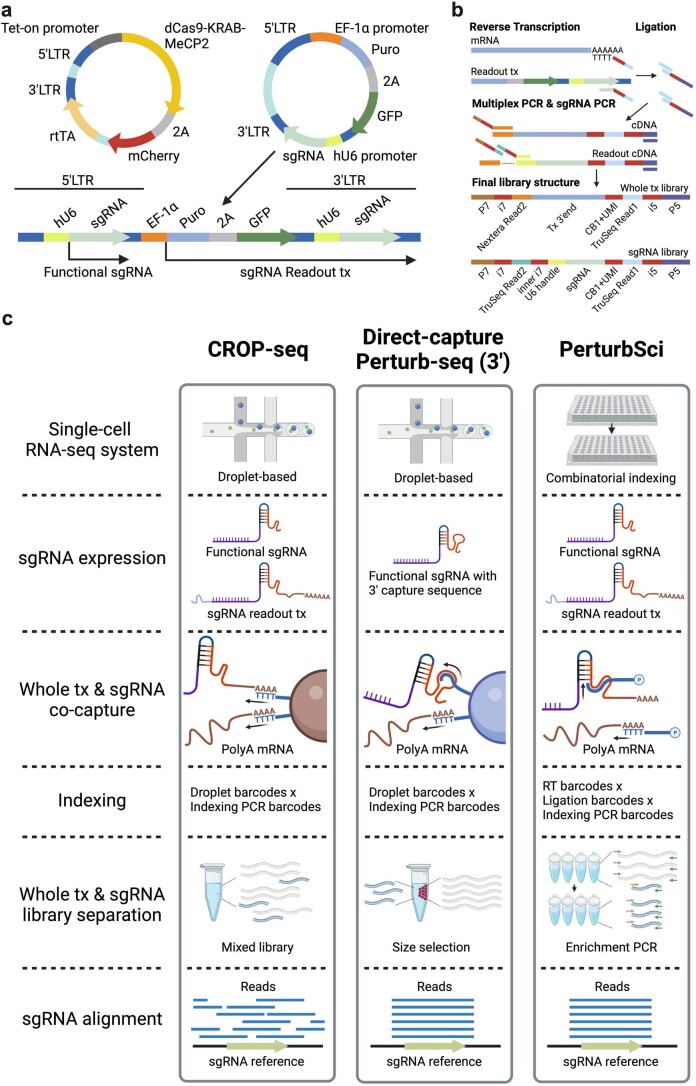
Extended Data Fig. 1. Scheme of plasmids and experiment procedures of PerturbSci.
a. The vector system used in PerturbSci for dCas9 and sgRNA expression. The expression of the enhanced CRISPRi silencer dCas9-KRAB-MeCP2 (ref. 9) was controlled by the tetracycline responsive (Tet-on) promoter. A GFP sequence was added to the original CROP-seq-opti plasmid6 as an indicator of successful sgRNA transduction and for the lentivirus titer measurement. The CROP-seq vector utilizes the self-replication mechanism of lentivirus during the integration for amplifying the sgRNA expression cassette. In this lentiviral plasmid, the sgRNA expression cassette replaced the U3 region of the 3′LTR5. During the lentiviral integration, the shortened 5′LTR of reverse-transcribed lentiviral genomic DNA was extended by using its 3′LTR as a template, and the sgRNA expression cassette is self-replicated to the 5′LTR62. The self-replicated sgRNA expression cassette at 5′LTR generates functional sgRNA for guiding dCas9, and the original expression cassette at 3′LTR is transcribed as a part of the Puro-GFP fusion transcript driven by the EF-1α promoter. b. The library preparation scheme and the final library structures of PerturbSci, including a scalable combinatorial indexing strategy with direct sgRNA capture and enrichment that reduced the library preparation cost, enhanced the sensitivity of the sgRNA capture compared to the original CROP-seq5, and avoided the extensive barcodes swapping detected in Perturb-seq6. c. A schematic comparison of chemistries between PerturbSci, CROP-seq5, and Direct-capture Perturb-seq7.