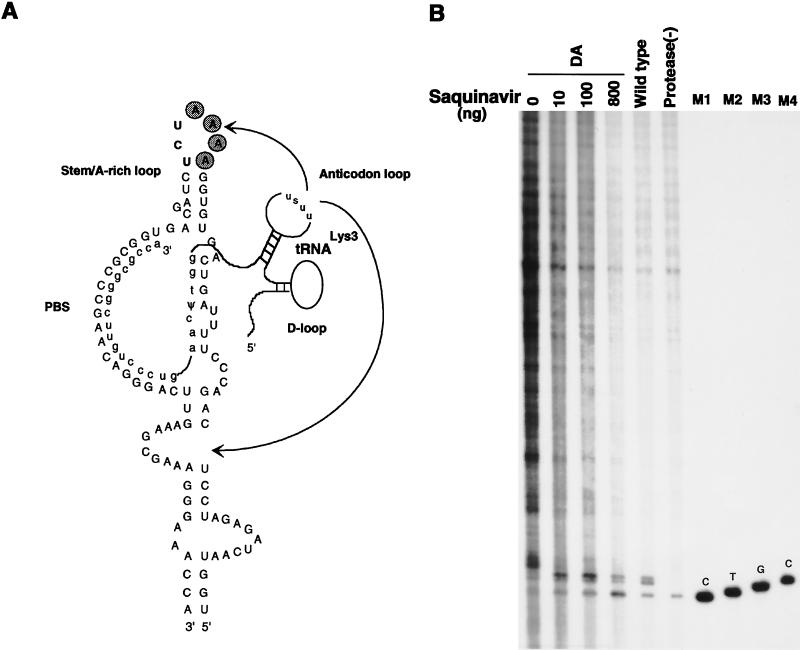
FIG. 6.
Effect of deletion of the four A’s in the A-rich loop on tRNA3Lys genomic placement and extension of tRNA3Lys by RT. (A) Proposed regions of base pairing between tRNA3Lys and the HIV-1 genome. This figure is modified from reference 3. In addition to the interaction between the 3′-terminal 18 nucleotides of tRNA3Lys and the PBS, other proposed interactions include ones between the tRNA3Lys anticodon loop and A-rich regions in the genome both upstream (20–22) and downstream (26) of the PBS (arrows), as well as a proposed interaction of the TΨC loop in the primer tRNA with a U5 region upstream of the PBS, which was initially proposed for avian retroviruses (1, 2). (B) Effect of A-rich loop deletion on tRNA3Lys placement and extension by RT. In vitro reverse transcription reactions were run as described in the Fig. 5 legend. Wild-type and protease(−) lanes represent total RNA isolated from wild-type and protease-negative virions, showing that only unextended tRNA3Lys is detected in protease-negative virions. The four DA lanes represent reactions with total RNA isolated from virus in which the four A’s of the A-rich loop have been deleted. The transfected cells, exposed to DNA for 15 h, were washed with fresh medium and were grown in increasing concentrations of the viral protease inhibitor Saquinovir for an additional 48 h before isolation of the virus. Lanes M1, M2, and M3, size markers generated from tRNA3Lys annealed to synthetic genomic RNA, as described in the legend to Fig. 5. Lane M4, size marker (4-base tRNA3Lys extension) generated by first extending tRNA3Lys 1 base with RT in the presence of [α-32P]dCTP and then adding dTTP, dGTP, and an excess of ddCTP before additional incubation.