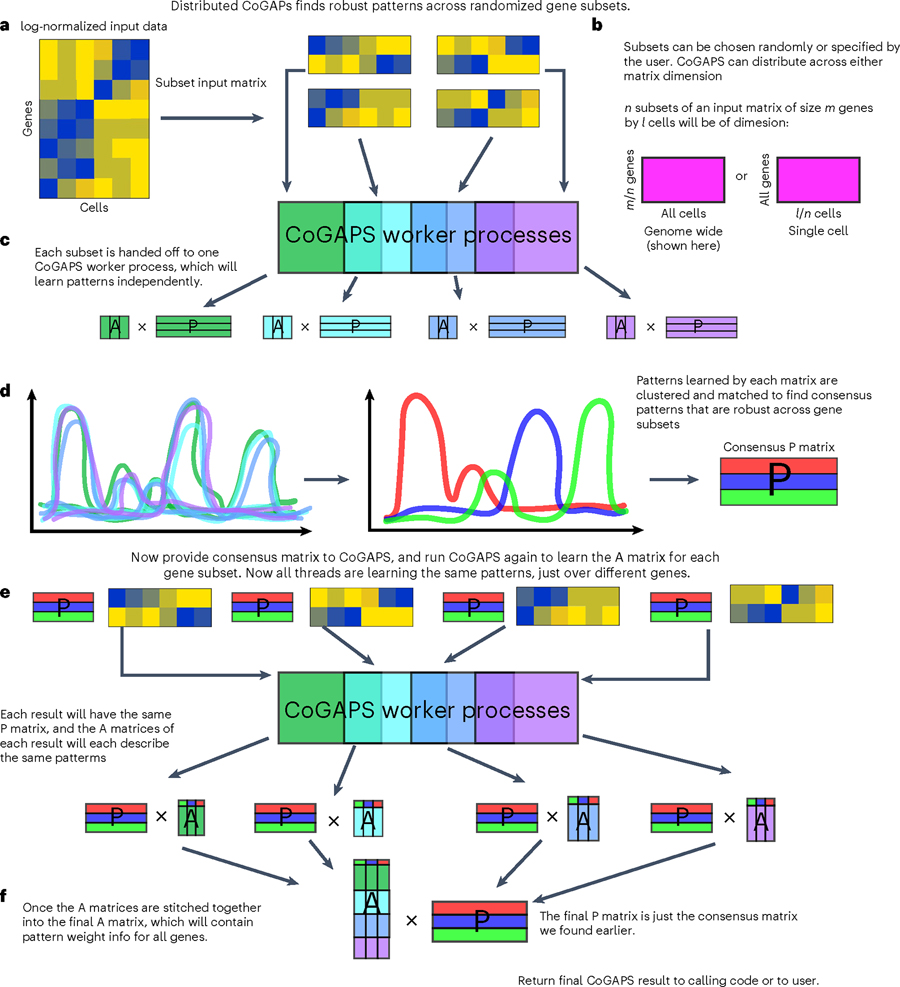
Fig. 3 |. Distributed CoGAPS finds robust patterns across randomized gene or sample subsets.
a, Subsetting is performed to break the input matrices into smaller components that can each be handed off to a worker process for NMF. b, Subsetting for parallelization can be performed across either matrix dimension. c, Each data subset yields its own NMF result. d, To identify the patterns that manifest themselves consistently across all NMF results, clustering is performed across all patterns returned by every thread, and a consensus matrix is generated from a process of matching cognate patterns. e, NMF is now run again on the same data subsets, this time with the consensus matrix provided as a ground truth from which the other matrix can be learned. This run is significantly faster than the first. f, Now that all threads have been forced to learn the same patterns, the portion of the NMF result that was not fixed can be stitched together to yield the final solution.