Figure 6. RNA-editing-dependent and -independent mechanisms of ADAR1 suppress aberrant dsRNA sensing.
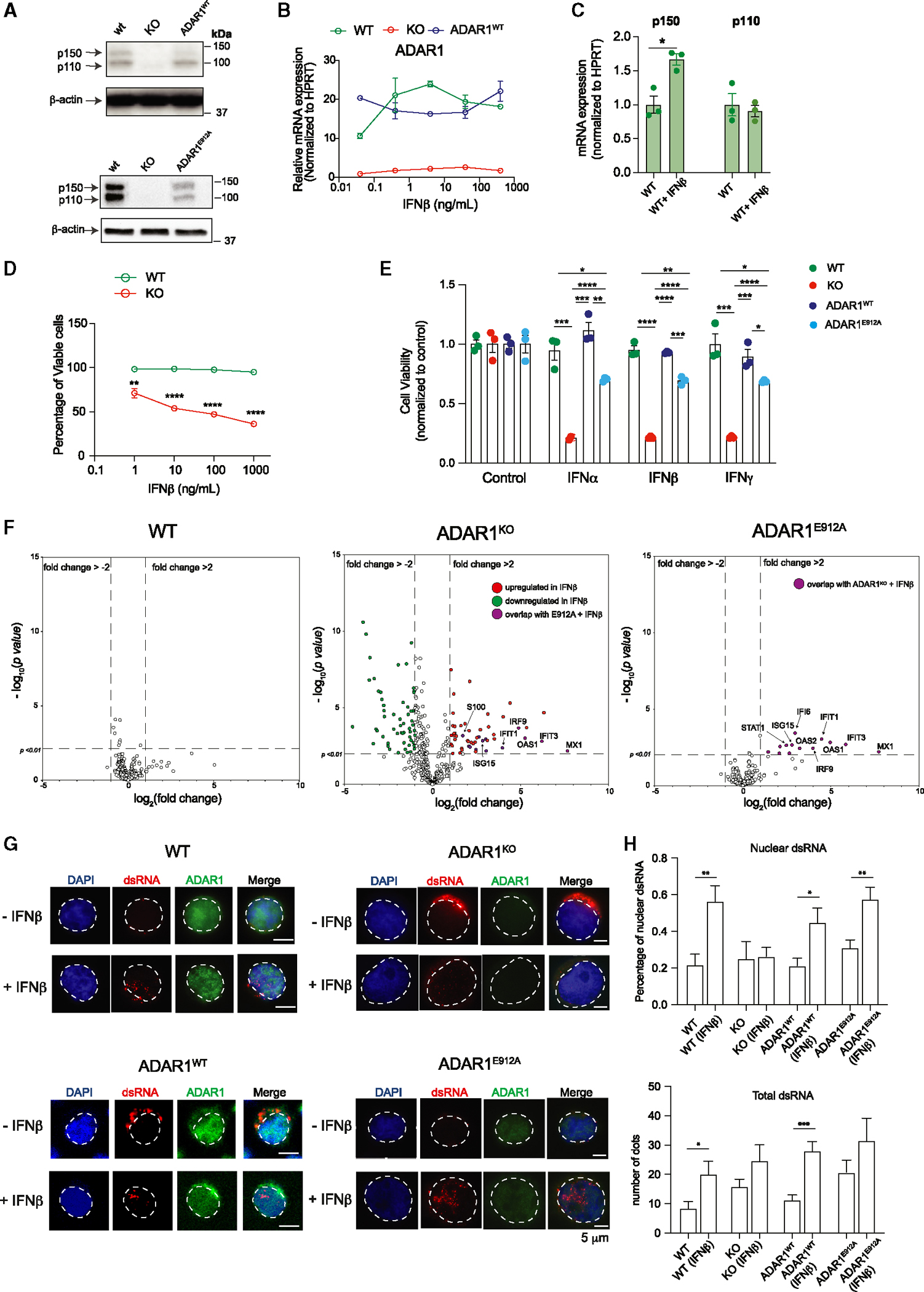
(A) Western blot showing ADAR1 expression in modified Jurkat T-ALL cell lines, including wild-type (WT), ADAR1-KO, re-expressed WT ADAR1 (ADAR1WT), and ADAR1E912A by lentiviral-overexpressing vector. β-Actin was used as loading control.
(B) Jurkat cells were stimulated with various concentrations of IFNβ, and the gene expression of ADAR1 was determined (n = 2 independent experiments).
(C) Quantification of ADAR1 p150 and p110 isoforms upon IFNβ treatment at 10 ng/mL for 24 h (n = 3 independent experiments).
(D) Cell viability was quantified in Jurkat cells upon IFNβ stimulation (n = 3 independent experiments).
(E) Cell viability was quantified in Jurkat cells treated with IFNα (0.1 ng/mL), IFNβ (1 ng/mL), or IFNγ (1 ng/mL) for 48 h (n = 3 independent experiments).
(F) NanoString analysis of gene expression in WT, ADAR1-KO, and ADAR1E912A-overexpressing Jurkat cells stimulated with IFNβ (1 ng/mL, 48 h) (n = 2 independent experiments).
(G) Immunofluorescent staining to detect the localization of dsRNA (J2 antibody) in WT, ADAR1-KO, ADAR1WT, and ADAR1E912A Jurkat cells stimulated with IFNβ (1 ng/mL, 24 h). Scale bars represent 5 μm.
(H) Quantification of total dsRNA dots and percentage of nuclear dsRNA from Jurkat cells treated with IFNβ (1 ng/mL), 24 h, 10 cells/condition.
*p < 0.05, **p < 0.01, and ****p < 0.0001, unpaired Student’s t test. Error bars represent mean with SEM in all graphs.
