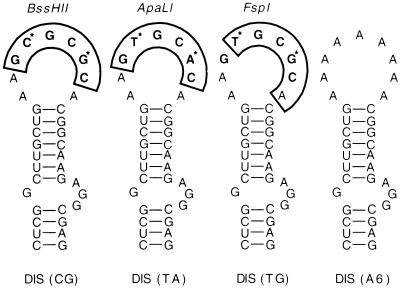
FIG. 1.
Predicted DIS RNA stem-loop structures of NL4-3 and NL4-3 DIS variants. The DIS mutants were generated by site-directed mutagenesis; for details, see Materials and Methods. The MFold program predicts the hairpin structures of all of the DIS variants; thus, alteration of the loop sequence minimally impacts stem structure. The palindromic motifs in the loop are in boldface, and restriction enzymes with corresponding cleavage sites (boxed) are shown above the hairpin loop. Variations at the second and fifth positions of the palindrome occurring in HIV-1 subtypes are marked (*). Phylogenetic association of DIS RNA elements of different HIV-1 strains is given in Table 1.