SUMMARY
The integrated stress response (ISR) is critical for cell survival under stress. In response to diverse environmental cues, eIF2α becomes phosphorylated, engendering a dramatic change in mRNA translation. The activation of ISR plays a pivotal role in the early embryogenesis, but the eIF2-dependent translational landscape in pluripotent embryonic stem cells (ESCs) is largely unexplored. We employ a multi-omics approach consisting of ribosome profiling, proteomics, and metabolomics in wild-type (eIF2α+/+) and phosphorylation-deficient mutant eIF2α (eIF2αA/A) mouse ESCs (mESCs) to investigate phosphorylated (p)-eIF2α-dependent translational control of naive pluripotency. We show a transient increase in p-eIF2α in the naive epiblast layer of E4.5 embryos. Absence of eIF2α phosphorylation engenders an exit from naive pluripotency following 2i (two chemical inhibitors of MEK1/2 and GSK3α/b) withdrawal. p-eIF2α controls translation of mRNAs encoding proteins that govern pluripotency, chromatin organization, and glutathione synthesis. Thus, p-eIF2α acts as a key regulator of the naive pluripotency gene regulatory network.
Graphical Abstract

In brief
mRNA translation is the final step of the central dogma of molecular biology. A key translational control mechanism is the phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2 (p-eIF2α). Amiri et al. identified the importance of translational control via p-eIF2α in the regulation of mouse ESCs.
INTRODUCTION
Mouse embryonic stem cells (mESCs) are derived from blastocysts and serve as an in vitro representation of the naive epiblast layer when cultured in a defined medium containing leukemia inhibitory factor (LIF) and two small-molecule kinase inhibitors, termed 2i: PD0325901 (PD, a mitogen-activated protein kinase [MAPK]/extracellular signal-regulated kinase [MEK] inhibitor) and CHIR99021 (CHIR, a glycogen synthase kinase-3 [GSK-3] inhibitor).1,2 Like epiblast progenitors, mESCs exhibit pluripotency and can differentiate into various cell types within the adult organism. Naive mESCs express pluripotency genes (e.g., Klf2, Klf4, Rex1), display global DNA hypomethylation, maintain two copies of active X chromosomes in female cells, and generate energy via oxidative phosphorylation and glycolysis.3 Although extensive research has focused on understanding the transcriptional and epigenetic regulators of naive pluripotency, studies on translational control are sparse. eIF4G2/DAP5/Nat1 and eIF4A2 are two examples of translational regulators of the naive state.4,5
The cellular proteome is predominantly shaped by mRNA translational control.6 Translational control is exerted primarily at the initiation step.7 Canonical translation initiation commences with the recruitment of the small ribosomal subunit (40S) to the cap structure (m7GpppN, where m represents a methyl group, and N denotes any nucleotide) at the mRNA 5′ end, followed by scanning of the 5′ UTR and recognition of the initiation codon, which is an AUG codon or a near-cognate codon that differs from AUG by a single nucleotide change. Initiation codon recognition requires the concerted action of translation initiation factors eIF1, eIF1A, eIF2, and eIF5 (a GTPase-activating protein for eIF2).8
The eukaryotic translation initiation factor 2 (eIF2) (consisting of α, β, and γ subunits) associates with GTP and the initiator tRNA (tRNAiMet) to form a ternary complex (TC) that directs the 40S ribosome to the initiation codon.9 Mammals express four eIF2α kinases (eIF2AKs) (heme-regulated inhibitor [HRI], eIF2AK1; protein kinase double-stranded RNA dependent [PKR], eIF2AK2; PKR-like endoplasmic reticulum kinase [PERK], eIF2AK3; and general control nonderepressible 2 [GCN2], eIF2AK4) which are activated by a bevy of extracellular and intracellular stresses.10 eIF2B is a guanine nucleotide exchange factor for eIF2 that exchanges GDP for GTP on eIF2. Phosphorylation of eIF2α converts eIF2 into an inhibitor of eIF2B,11 thereby depleting the TC pool and reducing protein synthesis. Thus, phosphorylated (p)-eIF2α is a pivotal regulator of the cell response to environmental cues and stress downstream of eIF2AKs in a pathway known as the integrated stress response (ISR). p-eIF2α is dephosphorylated by two heterodimeric eIF2α phosphatases: growth arrest and DNA damage-inducible protein 34 (GADD34) bound to protein phosphatase 1 (PP1) and constitutive repressor of eIF2α phosphorylation (CReP) bound to PP1.12,13 Although p-eIF2α curtails the translation of most cellular mRNAs, it paradoxically enhances the translation of a subset of mRNAs containing upstream open reading frames (uORFs) in their 5′ UTRs.8 The latter mRNAs encode proteins such as ATF4 (activating transcription factor 4), ATF5, CHOP, and GADD34 that mediate the unfolded protein response, oxidative stress response, and survival.14
The ISR pathway plays a pivotal role in the early development, as its dysregulation underlies numerous human developmental disorders, such as Wolcott-Rallison syndrome,15 MEHMO syndrome,16 Down syndrome,17 and vanishing white matter disease.18,19 While abrogation of eIF2α (eIF2S1 null mice) causes embryonic lethality prior to organogenesis (data from the International Mouse Phenotyping Consortium), eIF2αA/A mice develop to birth, but die postnatally due to severe hypoglycemia.20 Notably, loss of the two adaptor proteins of the eIF2α phosphatase complexes (Ppp1r15a−/−, Ppp1r15b−/−) engenders early embryonic lethality, which a single eIF2αS51A allele can rescue.21 These findings indicate that eIF2α and its dephosphorylation (but not its phosphorylation) is required for early development in non-stress conditions. However, eIF2α phosphorylation protects the embryo during stress conditions as GCN2−/− mice develop normally but exhibit severe developmental defects when leucine is absent in the maternal diet.22 Undifferentiated human and mouse ESCs exhibit elevated levels of p-eIF2α, which dramatically decreases upon differentiation.23 Here, we demonstrate that p-eIF2α transiently marks the naive epiblast cells of preimplantation mouse embryos. Furthermore, we discovered that the translational inhibition mediated by p-eIF2α operates synergistically with the inhibition of MEK1/2 and GSK3α/β to sustain the naive pluripotent state. The p-eIF2α controls the translation of mRNAs encoding proteins important for pluripotency, chromatin organization, and metabolism.
RESULTS
Transient enrichment of p-eIF2α in the naive epiblast layer of mouse pre-implantation embryo
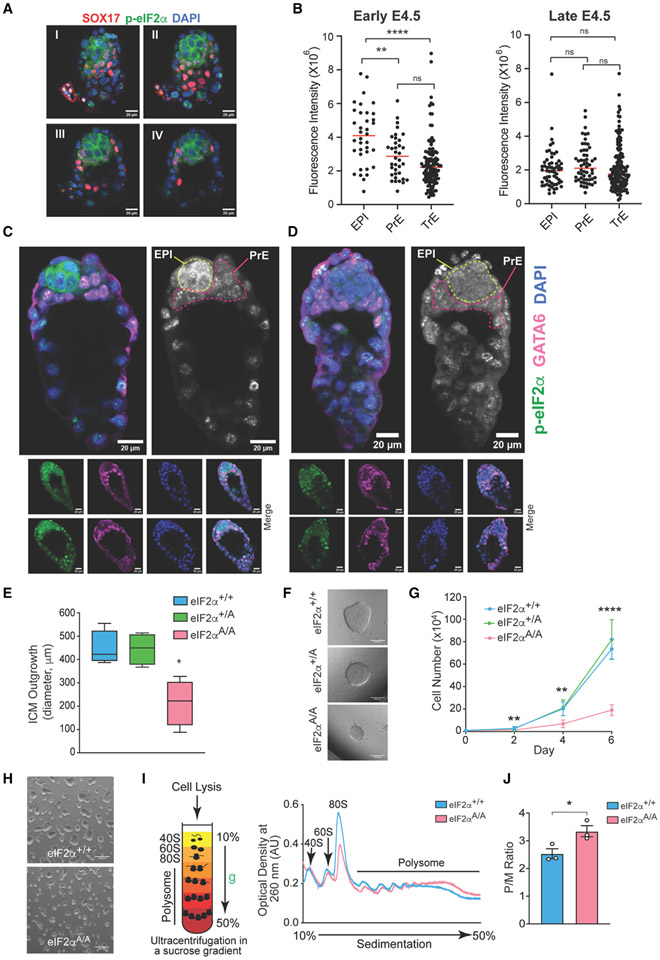
We first examined the distribution of p-eIF2α in vivo by immunostaining of mouse pre-implantation blastocyst (E4.5) (Figures 1A-1D). This is a critical developmental stage when the founders of all somatic cells are formed in the epiblast layer of the blastocyst. At E4.5, the inner cell mass (ICM) gives rise to two morphologically distinct lineages: primitive endoderm (PrE) (contributes to yolk sac and endoderm) and epiblast (EPI) (gives rise to the entire fetus and extraembryonic mesoderm). Co-staining for GATA6 (PrE marker) and p-eIF2α allows for accurate mapping of the distribution of p-eIF2α in the E4.5 embryo. At the early E4.5 stage, the level of p-eIF2α was significantly higher in the EPI layer than in the PrE and trophectoderm (TrE) layers (TrE; contributes to the placenta) (mean fluorescence intensity of 4.09 × 106 for EPI vs. 2.86 × 106 and 2.41 × 106 for PrE and TrE, respectively) (Figures 1A, 1B [left panel], and 1C). Notably, p-eIF2α is reduced in the EPI layer to the level seen in the PrE and TrE layers after ~3 h in late E4.5 embryos (Figures 1B [right panel] and 1D). The transient increase of p-eIF2α in the EPI of pre-implantation blastocysts suggests that p-eIF2α plays a role in regulating the naive state.
Figure 1. p-eIF2α transiently marks naive epiblast (EPI) lineages.
(A) Immunostaining analysis of an early E4.5 mouse embryo for p-eIF2α and Sox17 (primitive endoderm [PrE] marker). I, II, III, and IV represent serial optical sections of the same image. Scale bars, 20 μm.
(B) Quantification of p-eIF2α staining in each blastocyst lineage (EPI, PrE [GATA6 positive] and trophectoderm [TrE]) in early (left) and late (right) pre-implanting mouse embryos (E4.5). Each dot represents one cell from embryos (n = 5–6), **p< 0.01, ****p < 0.0001, ns, not significant, one-way ANOVA test with correction for multiple comparisons using Dunnett T3.
(C and D) Immunostaining of three early E4.5 and three late E4.5 embryos as described in (B), one embryo from each time point is magnified to show p-eIF2α staining of EPI in early E4.5, but not in late E4.5. Scale bars, 20 μm.
(E) Quantification of the ICM outgrowth diameters derived from culturing eIF2α+/+, eIF2α+/A, and eIF2αA/A blastocysts for 9 days (n = 4–5), *p < 0.05, one-way ANOVA test with correction for multiple comparisons using Dunnett’s T3 test.
(F) Representative image of ICM outgrowths described in (E). Scale bars, 200 μm.
(G) Cell proliferation assay for early passage (<6) eIF2α+/+, eIF2α+/A, and eIF2αA/A mESCs. **p < 0.01, ****p < 0.0001 (n = 5–6), two-way ANOVA test with correction for multiple comparisons using Tukey. Data presented as mean ± standard deviation (SD).
(H) Representative images of eIF2α+/+ and eIF2αA/A mESCs cultured in serum-free, feeder-free LIF/2i. Scale bars, 400 μm.
(I) Schematic representation of the fractionation of ribosomes using sucrose gradient. Cytoplasmic lysates of eIF2α+/+ and eIF2αA/A mESCs were loaded on a 10%–50% sucrose gradient, centrifuged (35,000 rpm for 2 h, 4°C), and UV absorbance at 260 nm was monitored continuously to detect 40S, 60S, 80S (monosomes [M]) and polysome fractions (P). Representative polysome profiles of eIF2α+/+ and eIF2αA/A mESCs cultured in serum-free, feeder-free LIF/2i are shown on the right.
(J) Quantification of P/M ratio of polysome profiles obtained from eIF2α+/+ and eIF2αA/A mESCs, *p < 0.05 (n = 3), t test with Welch correction. Data presented as mean ± standard error of mean (SEM).
p-eIF2α controls the naive state of pluripotency
Ppp1r15a−/−; Ppp1r15b−/− embryos (where p-eIF2α is maintained) die at the preimplantation stage and, when cultured in an mESC medium, fail to form a blastocyst cavity or hatch to establish an mESC line.21 To investigate the role of p-eIF2α in mESCs in pluripotency and lineage commitment, we derived mESCs from eIF2α+/+, eIF2α+/A, and eIF2αA/A littermate embryos20 (Figures S1A and S1B). Consistent with the original report,20 blastocysts produced eIF2αA/A mESCs at the expected Mendelian ratio (Figure S1A). However, ICM outgrowth of eIF2αA/A blastocysts was significantly smaller than those derived from eIF2α+/+ and eIF2α+/A blastocysts (average diameter of 215 μm for eIF2αA/A outgrowths vs. 447 and 445 μm for eIF2α+/+ and eIF2α+/A out-growths, respectively) (Figures 1E and 1F). p-eIF2α is elevated in mESCs cultured in the presence of LIF, BMP4, and GSK3β inhibitors, and LIF maintains p-eIF2α by CReP downregulation.23 Consequently, we asked whether mESCs cultured in the presence of LIF show growth differences that are dependent on the status of eIF2α phosphorylation. To exclude the contribution of serum or B27 and N2 supplements on p-eIF2α, we maintained mESCs in KnockOut Serum Replacement (KOSR). mESCs can be propagated continuously in bulk culture in the presence of LIF, 2i, and KOSR.24,25 Early-passage eIF2αA/A mESCs cultured in serum-free medium supplemented with LIF and 2i (LIF/2i) proliferated at a ~3.5-fold slower rate compared with eIF2α+/+ and eIF2α+/A mESCs (~1.9 × 105 cells on day 6 after plating for eIF2αAA mESCs vs. 7.4 × 105 and 8.2 × 105 cells for eIF2α+/+and eIF2α+/A mESCs, respectively) (Figures 1G and 1H). The similar growth of eIF2α+/+ and eIF2α+/A mESCs indicates that one eIF2αS51 allele is sufficient to rescue the slower proliferation rate of eIF2αA/A cells. Phosphorylation of eIF2α engenders a decrease in global translation.8 As expected, polysome profiling demonstrated an increase (~30%) in the polysome/monosome (P/M) ratio in eIF2αA/A mESCs compared with eIF2α+/+ cells (Figures 1I and 1J). ATF4 expression is critical for the proliferation of transformed cells26 and may explain the reduced proliferation of eIF2αA/A mESCs despite higher global translation. Surprisingly, alkaline phosphatase (AP) expression (measured by AP staining) or OCT3/4 and NANOG levels (measured by western blotting) were not reduced despite the slower proliferation of eIF2αA/A mESCs when compared with eIF2α+/+ and eIF2α+/A mESCs in LIF/2i medium (Figures S1C and S1D). To monitor the impact of p-eIF2α on the naive state, we removed 2i from the serum-free medium containing LIF (hereafter referred to as “LIF”27). On day 6 after 2i removal, eIF2αA/A mESCs displayed a >2-fold decrease in AP+ cells and in NANOG expression (Figures S1C, S1E, and S1F). This observation is consistent with a previous study in adult stem cells, which showed that, in the absence of eIF2α phosphorylation, the exit of muscle stem cells from the quiescence state is accelerated.28 Therefore, it is conceivable that, in the absence of 2i, p-eIF2α is required to maintain the naive pluripotent state and prevent spontaneous differentiation of mESCs.
To test the hypothesis, we examined whether inhibition of eIF2α phosphatases prevents mESC differentiation by using potent inhibitors of eIF2α phosphatases: sephin1 (S, GADD34: PP1 inhibitor)29 and raphin1 (R, CReP: PP1 inhibitor).30 As expected, the inhibitors blocked eIF2α dephosphorylation and rescued the expression of pluripotency factors, NANOG and OCT3/4, in the absence of 2i (Figures S1G and S1H). Notably, the effect of the SR inhibitors is solely due to the maintenance of p-eIF2α since eIF2αA/A mESCs did not respond to SR treatment. These data are consistent with previous reports that salubrinal (an inhibitor of eIF2α phosphatases) increases NANOG expression,23 whereas ISRIB (a compound that counteracts p-eIF2α by increasing eIF2B activity) exhibits an opposite effect on NANOG expression in breast cancer cell lines.31 Unlike the effect of phosphatase inhibitors, 2i rescues NANOG expression in both eIF2αA/A and eIF2α+/+ mESCs. These results indicate that p-eIF2α is required to maintain the naive state, which is overridden by the inhibition of MEK1/2 and GSK3α/β.
Translatome of eIF2αA/A mESCs
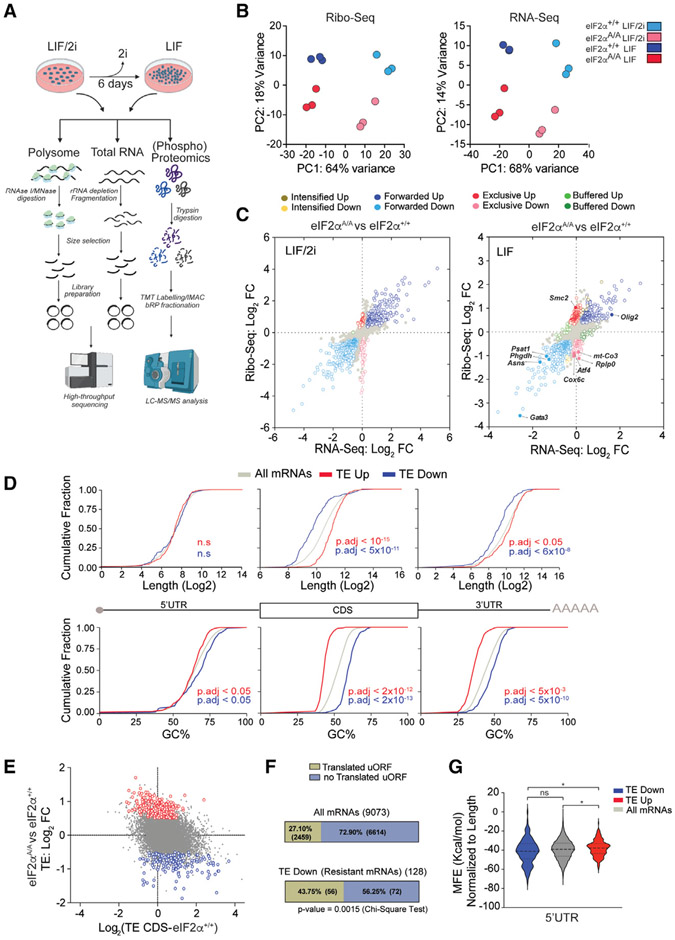
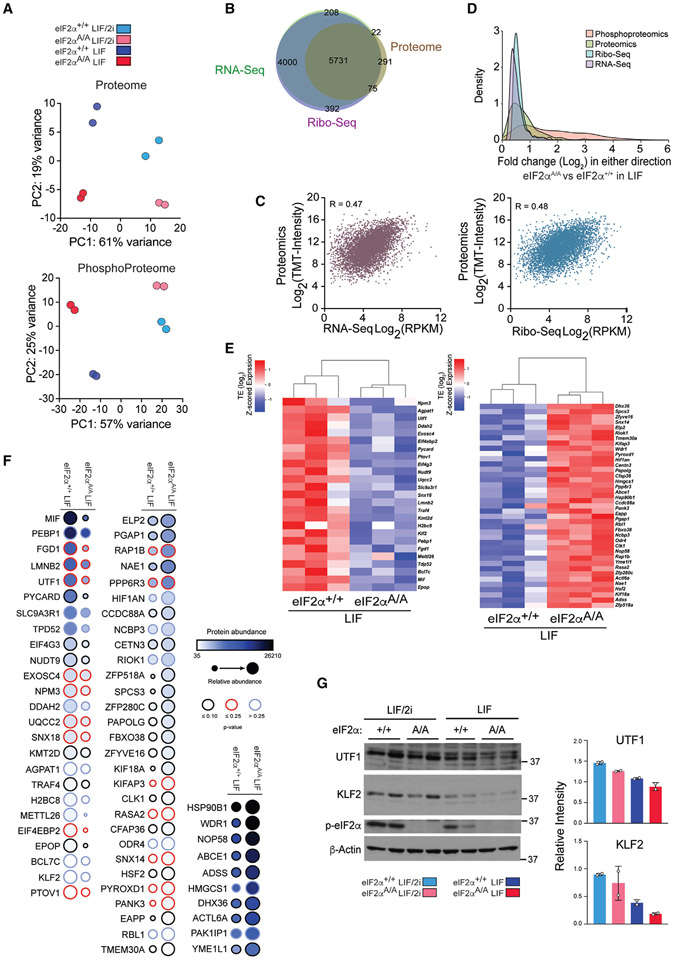
To delineate the mechanisms by which phosphorylation of eIF2α controls the naive state of pluripotency, we performed ribosome profiling (Ribo-seq)32 on eIF2αA/A and eIF2α+/+ mESCs grown under LIF/2i and LIF conditions (Figure 2A). In parallel, we monitored proteomic and phosphoproteomic changes using mass spectrometry (MS)-based proteomics (Figure 2A). Ribo-seq is a powerful sequencing-based technique that provides a singlenucleotide-resolution map for ribosome locations on mRNAs. It can also quantify the translation efficiency of mRNAs at the genome-wide level when coupled with RNA sequencing (RNA-seq).33,34 Two nucleases, ribonuclease I (RNase I) and micro-coccal nuclease (MNase), are often utilized to generate ribosome footprints. Both nucleases cleave the RNA at all four nucleotides.35 However, RNase I cleaves the mRNA at the exact edge of the ribosomes, which is preferred when sub-codon resolution is required, such as discovery of novel ORFs.36,37 In contrast, MNase does not cleave at the ribosome edge, likely because of its size difference and limited accessibility to the ribosome edge.36 Therefore, MNase maintains ribosome integrity by sparing rRNA, which renders it the nuclease of choice for assessing the translational efficiency of mRNAs.36,38 We used both nucleases to produce Ribo-seq libraries (Figure 2A). We obtained, on average, ~13M and ~19M mapped reads to the transcriptome for Ribo-seq and RNA-seq, respectively. MNase produced broader footprint length distributions with a peak centered at ~36 nucleotides (nt), whereas RNase I footprints peaked at ~29 nt with a narrower distribution (Figures S2A and S2C). As expected,35 metagene plots for both Ribo-seq datasets show the accumulation of footprints in the CDS of the mRNAs and triplet periodicity for RNAse I libraries (Figures S2B, S2D, and S2E). In agreement with a previous report,36 MNase libraries yielded a higher percentage of footprints associated with the 3′ UTR (Figure S2F) than RNase I-generated libraries. Principal-component analysis (PCA) and pairwise correlation of Ribo-seq and RNA-seq libraries revealed high reproducibility between biological replicates and well-defined separation of eIF2αA/A and eIF2α+/+ mESCs under LIF/2i and LIF conditions (Figures 2B, S2G, and S2H). We identified 10,198 and 9,961 highly expressed mRNAs (reads per kilobase per million mapped reads > 1) in Ribo-seq and RNA-seq, respectively (using the mRNA with longest CDS for each gene) (Table S1).
Figure 2. Translatome analysis of eIF2α+/+ and eIF2αA/A mESCs.
(A) Experimental design for preparation and sequencing of RNA-seq and Ribo-seq libraries, and phospho(proteomics) analysis of mESCs. This analysis was performed on biological replicates of eIF2α+/+ and eIF2αA/A mESCs cultured for 6 days in the serum-free medium containing LIF (1,000 U/mL) in the presence or absence of 2i. Image was created with https://biorender.com/.
(B) Principal-component analysis (PCA) plots of Ribo-seq (left) and RNA-seq (right) datasets separating each treatment and genotype. Each circle shows a biological replicate; different conditions and mESC genotypes are color coded.
(C) Scatterplot showing the association between RNA-seq and Ribo-seq fold change (FC) between eIF2αA/A and eIF2α+/+ mESCs under LIF/2i (left) or LIF (right) conditions. mRNAs with statistically significant changes (p-adj < 0.05 and log2 FC > 0.2 or log2 FC < −0.2) are highlighted with different colors according to the following categories: intensified up, increase in transcription, increase in translation efficiency (TE). Intensified down: decrease in transcription, decrease in TE. Forwarded up: increase in mRNA and RPF at the same rate, no change in TE. Forwarded down: decrease in mRNAand RPF at the same rate, no change inTE. Exclusive up: increase in RPF and TE, no change in mRNA. Exclusive down: decrease in RPF and TE, no change in mRNA. Buffered up: decrease in transcription/increase in TE. Buffered down: increase in transcription/decrease in TE.
(D) Cumulative frequency curves of length and GC% of different mRNA regions between translationally upregulated (TE up) and translationally downregulated (TE down) mRNAs in eIF2αA/Avs. eIF2α+/+ mESCs cultured in LIF medium. Adjusted p values are shown on each panel, n.s, not significant, one-way ANOVA test with correction for multiple comparisons using Games-Howell test.
(E) Correlation between TE of CDS of mRNAs in eIF2α+/+ cells and TE FC of eIF2αA/A vs. eIF2α+/+ cells in LIF medium.
(F) The percentage of translated and untranslated (not translated) AUG initiating uORFs in all mRNAs in the dataset and p-eIF2α-resistant (TE down in eIF2αA/A mESCs) mRNAs.
(G) Violin plots of minimum free energy (MFE) score normalized to 5′ UTR length for all mRNAs (gray), p-eIF2α-resistant (blue), and p-eIF2α-sensitive (TE up in eIF2αA/A mESCs; red) mRNAs. ns, not significant, *p < 0.05, one-way ANOVA test with correction for multiple comparisons using Dunnett T3.
We used the deltaTE (DTE) method39 to assess translation efficiency in eIF2α+/+ and eIF2αA/A mESCs in the presence or absence of 2i. DTE is an optimized version of DESeq240 for translation efficiency measurement that classifies genes based on changes in their Ribo-seq and RNA-seq results (Figure 2C; Table S2).39 Under the LIF/2i conditions, 44 mRNAs had significantly higher TE (p-adj < 0.05 and log2 fold change [FC] > 0.5) in eIF2αA/A compared with eIF2α+/+ mESCs (Figure 2C, left panel; intensified up + exclusive up + buffered up). Similarly, 53 mRNAs displayed significantly lower TE (p-adj < 0.05 and log2 FC < −0.5) in eIF2αA/A compared with eIF2α+/+ mESCs when cultured in LIF/2i medium (Figure 2C, left panel; intensified down + exclusive down + buffered down). Strikingly, in the absence of 2i (LIF conditions), the number of translationally regulated mRNAs increased 4-fold (Figure 2C, right panel; intensified up + exclusive up + buffered up = 232 mRNAs and intensified down + exclusive down + buffered down = 128 mRNAs). Notably, most TE changes under the LIF conditions belong to the “exclusive” category, where there is no change in mRNA levels while the Ribo-seq reads are significantly up- or downregulated. These data demonstrate that, in the absence of 2i, p-eIF2α-dependent translational control dominantly impacts gene expression.
General features of p-eIF2α-sensitive mRNAs
mRNA length, sequence, secondary structure, and the presence of uORFs play a pivotal role in controlling mRNA translation initiation.8 We analyzed translationally regulated mRNAs under the LIF conditions to explore mRNA features contributing to the differential translation response in eIF2αA/A mESCs. mRNAs with short CDS and 3′ UTRs and high GC content were resistant to translation inhibition by p-eIF2α (mean log2 length of CDS for TE down mRNAs, 9.83 vs. 11.07 for TE up mRNAs; mean GC% of CDS for TE down mRNAs, ~60% vs. ~43% for TE up mRNAs) (Figure 2D). Shorter mRNAs were reported to exhibit higher translation efficiency,41 and therefore should remain translationally active during inhibition of global translation. This was proposed by Lodish42-44 almost 50 years ago based on experiments in a rabbit reticulocyte translation extract that was subsequently supported in yeast models.42-44 While enrichment of short mRNAs in p-eIF2α-resistant mRNAs is consistent with Lodish’s model, enrichment of mRNAs with high GC content is not. However, two reports identified a strong correlation between lower GC content and accumulation of mRNAs in stress granules or P-bodies.45,46 p-eIF2α promotes stress granule formation, which may account for the enrichment of mRNAs with high GC content among p-eIF2α-resistant mRNAs. The TE FC represents the difference between TE in eIF2αA/A vs. TE in eIF2α+/+ mESCs. An inverse correlation exists between TE of CDS of mRNAs in eIF2α+/+ cells and TE FC of eIF2αA/A vs. eIF2α+/+ cells (Figure 2E). These data indicate that mRNAs with lower TE in eIF2αA/A mESCs are not contributing to the translation inhibition observed in eIF2α+/+ mESCs (Figure 2E). Next, we analyzed the presence of uORFs in TE down mRNAs in eIF2αA/A mESCs (hereinafter “p-eIF2α-resistant mRNAs”). Forty-three percent of p-eIF2α-resistant mRNAs possess at least one translated uORF, as detected by ribosome footprints in their 5′ UTR. In contrast, only 27% of mRNAs in the entire dataset contain a translated uORF (Figure 2F). Although the mere presence of translated uORFs does not determine resistance to p-eIF2α, our data demonstrate that translated uORFs are significantly enriched in p-eIF2α-resistant mRNAs. Consistent with a previous report,14 5′ UTRs of p-eIF2α-resistant mRNAs have a higher potential for secondary structure formation than p-eIF2α-sensitive mRNAs (Figure 2G).
p-eIF2α-dependent transcriptional reprogramming in mESCs upon 2i removal
Gene ontology (GO) analysis of translationally regulated mRNAs reveals that several transcription factors and genes involved in RNA polymerase II (RNAP II) functions (e.g., Tcf3, Atf4, Tcf7, and Esrra) are among the p-eIF2α-resistant mRNAs (Figure 3A, top panel). We found a better correlation between RNA-seq and Ribo-seq in eIF2α+/+ compared with eIF2αA/A mESCs (R = 0.91 for eIF2α+/+ mESCs and R = 0.88 for eIF2αA/A mESCs) (Figure S3A), suggesting a higher dependency of translation rates on mRNA abundance in eIF2α+/+ mESCs. Consistent with a strong impact of translation regulation, RNA-seq FC was less correlated with Ribo-seq FC when comparing eIF2α+/+ with eIF2αA/A cells (R = 0.64 in LIF/2i and R = 0.57 in LIF) (Figure S3B).
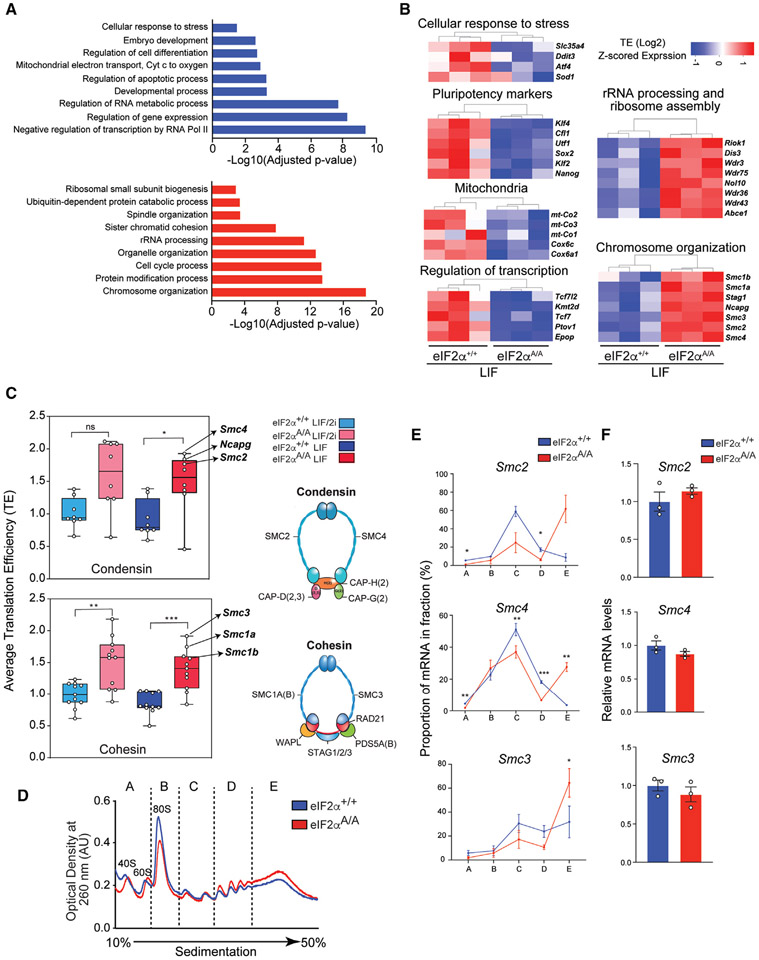
Figure 3. p-eIF2α controls the translation of mRNAs encoding key cellular processes.
(A) Gene ontology (GO) analysis for translationally downregulated (top panel) and upregulated (bottom panel) mRNAs in eIF2αA/A compared with eIF2α+/+ mESCs under LIF conditions. p values were corrected for multiple comparisons using the Benjamini-Hochberg method. mRNAs with p-adj < 0.1 and −0.5 > log2 FC>0.5 are considered for this analysis; up mRNAs, n = 408; down mRNAs, n = 259.
(B) Heatmaps highlight translationally upregulated and downregulated genes (eIF2αA/A compared with eIF2α+/+ mESCs under LIF conditions) that define representative functional categories. Values show log2 transformed z-scored TE.
(C) Average TE of components of condensin and cohesin complexes. The complete list of components and the corresponding TE are provided in Table S6. *p < 0.05, **p < 0.01, ***p < 0.001, ns, not significant. One-way ANOVA test with correction for multiple comparisons using Tukey.
(D) eIF2α+/+ and eIF2αA/A mESCs were cultured under LIF conditions. Sub-polysome (A and B) and polysome (C, D, and E) fractions were obtained by ultra-centrifugation through 10%–50% sucrose gradients.
(E) The percentage of mRNAs in each fraction was quantified by RT-qPCR. *p < 0.05, **p < 0.01, ***p < 0.001. (n = 3), two-way ANOVA test with correction for multiple comparisons using the Benjamini-Hochberg method. Data presented as mean ± SEM.
(F) The total mRNA levels of the genes assessed in (E) were quantified using RT-qPCR of the input. (n = 3), t test with Welch correction. Data presented as mean ± SEM.
Among the GO groups that are transcriptionally downregulated in eIF2αA/A cells (Figure S3C, top panel) are mRNAs encoding proteins linked to developmental processes, vascular development, MAPK pathway, and several known ATF4 targets such as mRNAs encoding proteins of glutathione metabolism47 (see below), L-serine metabolism,48 and tRNA aminoacylation.49 Whereas developmental-related mRNAs are transcriptionally downregulated in eIF2αA/A mESCs, mRNAs whose proteins participate in neuron differentiation, neuron projection morphogenesis, and gliogenesis are upregulated (Figure S3C, bottom panel).
In response to 2i withdrawal, eIF2α+/+ and eIF2αA/A mESCs display a drastic transcriptome reprogramming (Figure S3D). Large numbers of genes (2,282 mRNAs in eIF2α+/+ mESCs and 1,265 mRNAs in eIF2αA/A mESCs) show similar changes in RNA-seq and Ribo-seq (at the same rate). Therefore, no significant change in TE was detected (forwarded up or down categories, p-adj < 0.05) (Figure S3D). Consistent with the removal of 2i, several downstream targets of the ERK pathway, such as DUSP6, DUSP4, and MYC, are induced as determined by Ribo-seq and proteomic analysis (Figures S3E and S5C). Several naive pluripotency markers (e.g., KLF2, ESRRB, ZFP42, TBX3, and TFCP2L1) are downregulated with concurrent upregulation of primed pluripotency markers,27 such as OTX2, DNMT3B, and DNMT3A (Figures S3E and S5C).
p-eIF2α regulates the translation of mRNAs that play crucial roles in development, mitochondrial energy metabolism, and chromatin structure
In addition to transcriptional regulators (e.g., Tcf7l2, Kmt2d, Tcf7, Ptov1, and Epop), GO analysis of p-eIF2α-resistant mRNAs showed enrichment for those involved in embryo development and differentiation (e.g., Klf4, Cfl1, and Utf1), glutathione synthesis (e.g., Gpx1, Gstm1, and Slc25a39), mitochondrial electron transport (e.g., Mt-co2, Mt-co3, and Cox6c) and cellular response to stress (e.g., Slc35a4, Ddit3, Atf4, and Sod1)(figures 3A[top panel] and 3B). In contrast, translationally activated mRNAs in eIF2αA/A mESCs (hereinafter called “p-eIF2α-sensitive mRNAs”) were enriched for mRNAs involved in spindle organization, sister chromatid cohesion, and chromosome organization (e.g., Smc1b, Ncapg, and Smc4), rRNA processing, ribosome biogenesis, and ubiquitin-dependent processes (Figures 3A [bottom panel] and 3B). Consistent with the finding that phosphorylation of eIF2α controls the naive state, TE of several naive markers such as Klf2, Klf4, and Nanog were decreased in eIF2αA/A compared with eIF2α+/+ mESCs (Figure 3B). Polysome profiling was performed to directly examine whether eIF2α phosphorylation controls the translation initiation of identified targets. mRNAs with faster translation initiation rates are associated with heavy polysomes, whereas poorly translated mRNAs are associated with light polysomes in a density gradient. Significantly, Utf1, Atf4, and Epop mRNAs were associated with heavier polysomes in the eIF2α+/+ mESCs (fractions D and E) compared with eIF2αA/A mESCs (fraction C) (Figures S3F, 4B, and 4D). In sharp contrast, Smc2, Smc4, and Smc3 mRNAs were associated with heavier polysomes in eIF2αA/A (fraction E) in comparison with the eIF2α+/+ mESCs (fraction C) (Figure 3E). Importantly, no differences were found in total mRNA levels between eIF2α+/+ and eIF2αA/A mESCs (Figure 3F). Taken together, these results validate our Ribo-seq analysis.
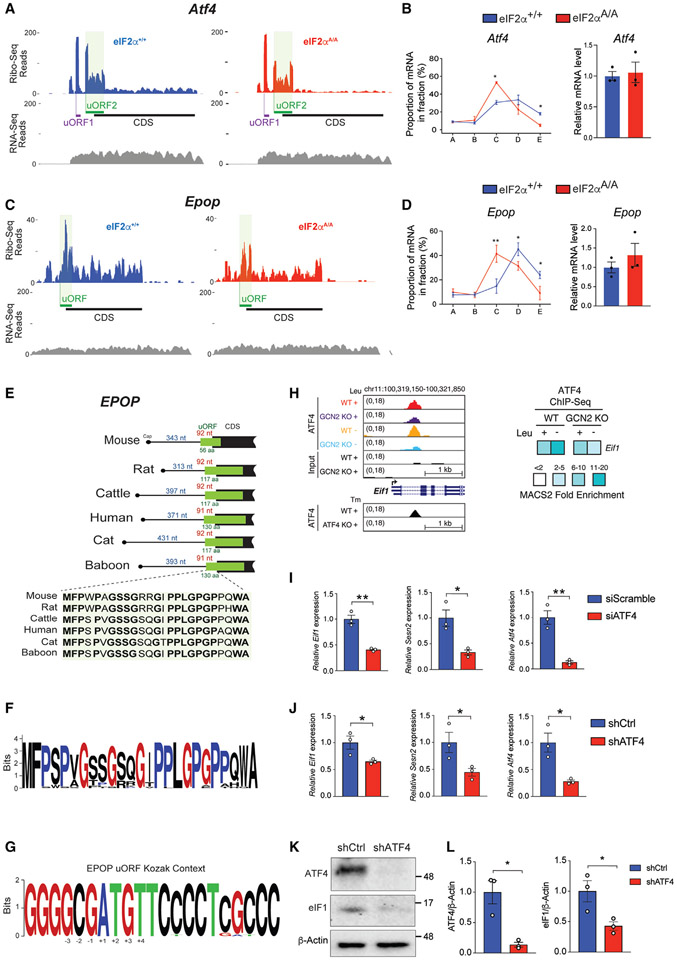
Figure 4. Identification of p-eIF2α-dependent regulatory uORFs.
(A) Ribosome (Ribo-seq) and RNA (RNA-seq) coverage plots for Atf4 mRNA in eIF2α+/+ and eIF2αA/A mESCs under LIF conditions.
(B) The percentage of Atf4 mRNAs in each fraction of polysome profiles (left) and total input mRNA level (right) of eIF2α+/+ and eIF2αA/A mESCs cultured in LIF medium were quantified by RT-qPCR. *p < 0.05, (n = 3), two-way ANOVA test with correction for multiple comparisons using the Benjamini-Hochberg method. Data presented as mean ± SEM.
(C) Ribo-seq and RNA-seq coverage plot of Epop in eIF2α+/+ and eIF2αA/A mESCs under LIF conditions.
(D) The percentage of Epop mRNAs in each fraction of polysome profiles (left) and total input mRNA level (right) of eIF2α+/+ and eIF2αA/A mESCs cultured in LIF medium were quantified by RT-qPCR. *p < 0.05, **p < 0.01, (n = 3), two-way ANOVA test with correction for multiple comparisons using the Benjamini-Hochberg method. Data presented as mean ± SEM.
(E) Position and amino acid conservation of Epop uORF-encoded protein in several mammals. The number of nucleotides (nt) between the cap and the start of uORF is indicated in blue for each species. The distance of the uORF start site to the CDS start site, and the length of the uORF-encoded protein are indicated in red and green, respectively, for each species.
(F) Amino acid conservation of N-terminal region of EPOP uORF-encoded peptide in mammalian species.
(G) Conservation of nucleotides flanking the start site of Epop uORF in mammalian species.
(H) Left: UCSC Genome browser views for ATF4 binding events in GCN2+/+ wild-type (WT) and GCN2−/− knockout (KO) cells that were exposed to control (+Leu) or leucine-deficient (−Leu) medium. ATF4 binding profiles in ATF4+/+ (WT) and ATF4−/− (KO) cells treated with tunicamycin (Tm) (GSE35681) are shown below the panel. Right: the heatmap shows fold enrichment for ATF4 ChIP-seq target eIF1.
(I and J) RT-qPCR analysis of N2A cells transduced with the indicated siRNA or shRNAs. Values were normalized to β-actin level. *p < 0.05, **p < 0.01. Data are presented as mean ± SEM (n = 3) t test.
(K and L) Western blot analysis and quantification of ATF4 and eIF1 in N2A cells transduced with shCtrl or shRNA against ATF4. Values were normalized to β-actin level. Results are presented as mean ± SEM (n = 3). *p < 0.05, t test.
p-eIF2α coordinates the translation of mRNAs encoding core components of cohesin and condensin in mESCs
We uncovered several components of multi-protein complexes or functionally related proteins that display concomitant sensitivity to eIF2α activity (Figure 3B). This finding is in accord with extensive data highlighting the precise proportional synthesis of multi-protein complex components (proportional synthesis).50 GO analysis of p-eIF2α-sensitive genes demonstrated enrichment for genes involved in chromatin structure and segregation (Figure 3B). Notably, multiple components of condensin and cohesin were among the p-eIF2α-sensitive mRNAs (Figures 3B and 3C). Consistent with Ribo-seq results, the mRNAs for condensin/cohesin components (Smc2, Smc4, and Smc3) were shifted to heavier polysome fractions without significant change in mRNA levels in eIF2αA/A mESCs, coinciding with a significant increase in the corresponding protein levels (Figures 3D-3F and S3G). Surprisingly, the high levels of p-eIF2α in wild-type mESCs cultured under LIF/2i conditions do not induce ATF4 expression (Figures S1G, 5G and S3G). This might be explained by the repressing effect of GSK3β on eIF2B.51,52 While the presence of GSK3β inhibitor in the LIF/2i medium blocks the inhibitory effect of GSK3β on eIF2B, 2i removal allows GSK3β inhibition of eIF2B, thereby leading to an increase in ATF4 expression. Notably, in addition to cohesin and condensin complexes, Ribo-seq revealed p-eIF2α-dependent tuning of translation of subunits of the THO complex53 (Figure S3H).
Figure 5. Proteomics complements ribosome profiling-based quantification of protein synthesis.
(A) Principal-component analysis (PCA) plots of the proteome (top) and phosphoproteome (bottom) datasets, separating each treatment and genotype. Each circle shows a biological replicate, and different conditions and mESC genotypes are color coded.
(B) Venn diagram illustrating the mRNAs identified by Ribo-seq, RNA-seq, and proteomics. The mRNAs in Ribo-seq (reads per kilobase per million mapped reads [RPKM] >1), RNA-seq (RPKM >1), and proteomics (S/N ratio >250) yielding 5,731 common gene products.
(C) Scatterplots showing the correlation between proteomics with RNA-seq (left) and Ribo-seq (right). R values are Pearson correlation coefficient.
(D) The distribution of the magnitude of changes in RNA-seq, Ribo-seq, proteomics, and phosphoproteomics datasets.
(E) Heatmap displaying a subset of translationally upregulated and downregulated genes (eIF2αA/A compared with eIF2α+/+ mESCs under LIF conditions) with similar changes in proteomics analysis. Values show log transformed z-scored TE.
(F) Dot plot showing the proteins with similar changes in protein level as in TE. Protein abundance is the average S/N ratio of TMT intensity of indicated proteins from two replicates. Color-coded border lines indicate p values.
(G) Western blot analysis and quantification of indicated proteins in two independent clones of eIF2α+/+ and eIF2αA/A mESCs cultured under the serum-free LIF/2i and LIF conditions for 6 days. Values were normalized to β-actin level. Data are presented as mean ± SD (n = 2). t test with Welch’s correction.
The pervasive presence of uORFs in p-eIF2α-resistant and -sensitive mRNAs
Forty-three percent of p-eIF2α-resistant mRNAs possess at least one translated uORF in their 5′ UTRs (Figure 2F). Generally, uORFs reduce the translation of downstream CDS.8 Distinct features of uORFs influence the recognition of downstream initiation codons by modulating leaky scanning and/or re-initiation. Important uORF features include the sequence context of AUG or near-cognate AUG start codons, the number of uORFs, length, and distance from the CDS.8 We first examined the Atf4 mRNA, arguably the best-characterized translational target of p-eIF2α in mammals.54 As expected, Ribo-seq only showed background levels of reads across the Atf4 CDS in eIF2αA/A mESCs, whereas footprints significantly increased across Atf4 CDS in eIF2α+/+ mESCs, indicating better translation because of eIF2α phosphorylation (Figure 4A). Atf4 contains two uORFs that exert opposite effects on the translation of the Atf4 CDS. uORF1 stimulates Atf4 translation54 while the inhibitory uORF2 extends into the CDS. The efficient translation of a uORF and inefficient translation of the CDS are predictors of p-eIF2α-resistant genes.14 Accordingly, footprint density on Atf4 uORF2 was higher than downstream CDS. Polysome profiling demonstrated that the Atf4 mRNA is associated with heavier polysome in eIF2α+/+ mESCs compared with eIF2αA/A mESCs without a significant difference in Atf4 mRNA level (Figure 4B). We identified an out-of-frame overlapping uORF on the mRNA of another key transcriptional regulator Epop (Elongin BC And polycomb repressive complex 2 [PRC2]-associated protein),55,56 which displays efficient translation of its uORF and inefficient translation of its CDS (Figures 4C and S4A-S4C). Polysome profiling confirmed elevated translation of Epop mRNAs in eIF2α+/+ mESCs compared with eIF2αA/A mESCs, without a difference in Epop mRNA, supporting a translational control mechanism (Figure 4D). The mouse Epop uORF encodes a 56 amino acid long polypeptide, which is highly conserved at the first 25 amino acids across vertebrates (Figures 4E-4G). EPOP is expressed only in the inner cell mass and plays a critical role in repressing PRC2 targets.56 The significance of the cis-regulatory elements in the Epop 5′ UTR on EPOP expression and its impact on PRC2 function remains to be determined. Other examples of p-eIF2α-resistant mRNAs with prominent uORF peaks are pluripotency factor Nanog and a nitric oxide regulator Ddah2 (dimethylarginine dimethylaminohydrolase 2) (Figures S4D and S4E). The uORF peak in Ddah2 mRNA overlaps with a uORF that contains a CUG start codon (Figure S4E). It is not clear why the footprints of Nanog uORF, unlike other p-eIF2α-resistant genes, did not display a strong peak compared with the CDS.
eIF1 and eIF5 are two other important initiation factors mediating the stringency of start codon selection with opposing roles.57,58 eIF1 increases start codon selection stringency, promoting the bypass of weak start sites, while eIF5 promotes initiation at weak start sites.57,58 One plausible mechanism for bypassing a single inhibitory uORF in p-eIF2α-resistant mRNAs is by ATF4-dependent activation of eIF1 gene transcription. Published ChIP-Seq data from our lab59 and others49 show that ATF4 directly binds to the promoter of eIF1 (Figure 4H). Consistently, the knockdown of ATF4 in N2A cells resulted in lower expression of eIF1 at the mRNA and protein levels (Figures 4I-4L; Sesn2, a well-established target of ATF4, was used as a positive control). Thus, start codon stringency selection could be mediated by eIF2α phosphorylation via ATF4/eIF1. Notably, the presence of translated uORFs is not restricted to eIF2α-resistant mRNAs and is also present in p-eIF2α-sensitive mRNAs, such as Zfp518a, Smc2, and Ncapg (Figures S4F and S4G).
MS-based proteomics and phosphoproteomics of the p-eIF2α-dependent proteome
While Ribo-seq provides a valuable proxy for assessing ribosome positions, indicating translating, or stalled non-translating ribosomes, it does not provide direct information on proteome composition. To study the impact of p-eIF2α on the proteome, we performed MS on eIF2α+/+ and eIF2αA/A mESCs. PCA of MS-based proteomics data showed high reproducibility between biological replicates and well-defined separation of LIF/2i and LIF, as well as eIF2αA/A and eIF2α+/+ mESC conditions (Figure 5A, top panel). A total of 6,164 qualified proteins was identified by MS analysis (see STAR Methods; Table S3). Integration of Ribo-seq, RNA-seq, and proteome data yielded 5,731 genes commonly detected by the three methods (Figure 5B). The Ribo-seq and RNA-seq analysis showed similar correlations with the proteome (Pearson correlation coefficient [R] = 0.47 for RNA-seq vs. 0.48 for Ribo-seq) (Figure 5C). Comparing each genotype in the presence and absence of 2i revealed that there is a higher correlation between RNA-seq and MS-based proteomics in eIF2αA/A mESCs compared with eIF2α+/+ mESCs (R = 0.43 for eIF2α+/+ mESCs and R = 0.50 for eIF2αA/A mESCs) (Figures S3A and S5A), in support of the notion that the effect of p-eIF2α-dependent transcriptional reprogramming on the proteome is dampened upon eIF2α phosphorylation. Several translationally regulated targets (64 targets) identified in Ribo-seq displayed similar changes in the proteomic analysis (Figures 5E and 5F; Table S4). In agreement with the Ribo-seq analysis, the amounts of EPOP and DDAH2 proteins were reduced in eIF2αA/A mESCs (Figures 4C, 4D, S4E, and 5F). Pluripotency factors UTF1 and KLF2 were also lower in eIF2αA/A mESCs (Figures 5E-5G).
Next, we examined the phosphoproteome in eIF2α+/+ versus eIF2αA/A mESCs (Figure S5B). PCA analysis demonstrated that 2i withdrawal had a greater impact on the phosphoproteome than on the proteome, as indicated by the larger separation of repeats in response to 2i removal (Figure 5A, bottom panel). The broader distribution of the phosphoproteome agrees with a previous finding that the phosphoproteome is more dynamic than the proteome and transcriptome during exit from the naive state (Figure 5D).60 Consistent with the removal of 2i and activation of ERK and GSK3 signaling pathways, levels of p-ERK and p-GSK3 increased (~4-fold) upon 2i withdrawal (Figure S5D; Table S4), which caused increased expression of several downstream targets, such as DUSP6, DUSP4, and MYC, at both the RNA and protein levels (Figures S3E and S5C). A phosphoproteome analysis revealed several eIF2α-dependent phosphorylated proteins that are involved in lineage commitment (Sox2 S39)61 or translational control (eIF4B S406/409) (Figure S5E). It is conceivable that feedback loops and crosstalk between ISR and other kinases, such as mTORC1, contribute to some of the phosphorylation changes.
p-eIF2α controls glutathione synthesis
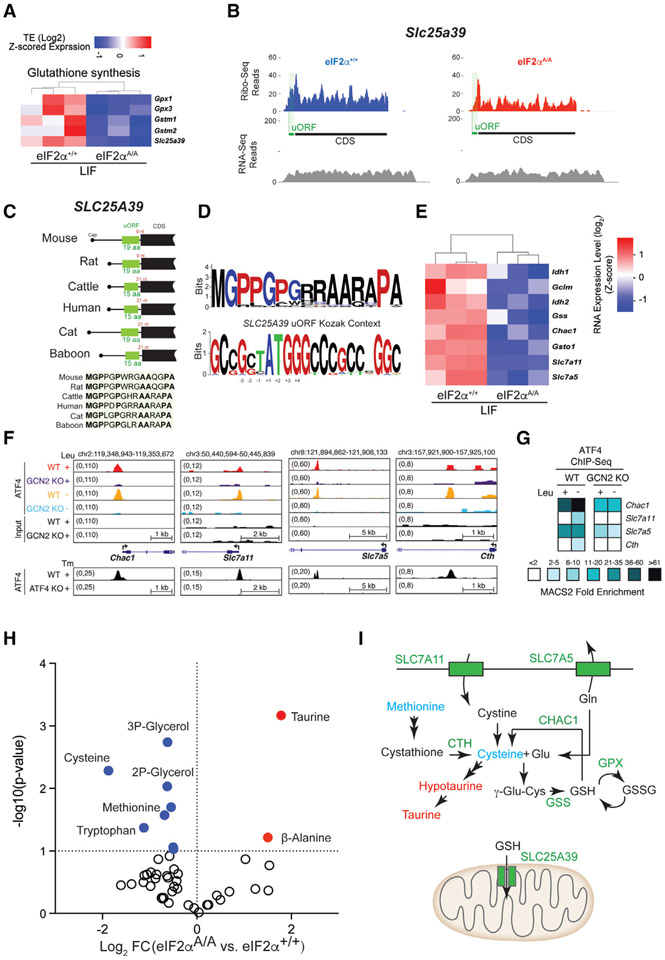
Among the functionally related p-eIF2α-resistance genes, a salient enrichment for mRNAs whose encoded proteins are involved in glutathione synthesis is evident (Figures 6A, S6A, and S6B). This subset of mRNAs includes Gpx1 and 3 (glutathione peroxidase 1 and 3), Gstm1 and 2 (glutathione S-transferase mu 1 and 2), and Slc25a39 (solute carrier family 25 member 39). Glutathione (GSH) is a small tripeptide molecule (L-gamma-glutamyl-L-cysteinyl-glycine) that has essential roles in numerous biological processes, including oxidative metabolism, protein synthesis, DNA synthesis, and metabolism.62 SLC25A39 is an indispensable component of the mitochondrial GSH-import machinery.63,64 Slc25a39 mRNA displays a prominent footprint corresponding to a uORF which terminates in mammals 9 to 21 nt upstream of the CDS (Figures 6B and 6C). Similar to other mRNAs described above containing inhibitory uORFs (Figure 4), Slc25a39 displays many reads of the uORF and few of the CDS (Figure 6B). Mouse Slc25a39 uORF encodes a 19 amino acid long polypeptide that is evolutionarily conserved across vertebrates (Figures 6C, 6D, S6D, and S6E). It contains two contiguous prolines and a Pro-Pro-Gly sequence identified in other inhibitory uORFs such as Gadd34 uORF2.65 SLC25A39 belongs to the SLC25A family of mitochondrial small-molecule carriers.66 Two additional members of this family, Ucp2 (uncoupling protein 2; Slc25a8; a mitochondrial anion carrier) and Slc25a19 (a thiamine pyrophosphate transporter),67 are among the p-eIF2α-resistance mRNAs that possess uORFs (Figure S6C). A recent study reported that ATF4 stimulates cystine uptake through transcriptional activation of the cystine-glutamate antiporter Slc7a11 gene.47 Accordingly, Slc7a11 mRNA amounts were significantly increased (~5-fold) in eIF2α+/+ compared with eIF2αA/A cells (Figure 6E). ChIP-seq data49,59 show that ATF4 binds to the promoter of Slc7a11 downstream of the ISR pathway (Figures 6F and 6G). Metabolomic analysis of eIF2α+/+ and eIF2αA/A ESCs revealed that cysteine levels were significantly decreased in the absence of eIF2α phosphorylation (Figure 6H), highlighting the importance of Slc7a11 in cystine uptake. Several other genes involved in glutathione synthesis were transcriptionally repressed in eIF2αA/A cells, and ChIP-seq analysis confirmed ATF4 binding sites in their promoters (Figures 6E-6G). Taken together, the data show that p-eIF2α controls multiple components of the glutathione metabolic pathway at both the translational and transcriptional levels (Figure 6I). This finding adds to the previously documented role of ATF4 in controlling the expression of genes involved in glutathione biosynthesis.68,69
Figure 6. p-eIF2α regulates the expression of mRNAs encoding proteins involved in glutathione synthesis.
(A) Heatmap plot highlighting translationally downregulated mRNA involved in glutathione synthesis (eIF2αA/A compared with eIF2α+/+ mESCs under LIF conditions). Values are log-transformed z-scored TE.
(B) Ribo-seq and RNA-seq coverage plot of Slc25a39 in eIF2α+/+ and eIF2αA/A mESCs under LIF conditions.
(C) Position and amino acid conservation of Slc25a39 uORF-encoded protein in several mammals. The distance of the uORF stop codon to the CDS start codon (in red) and the length of the uORF-encoded peptide (in green) have been shown for each species.
(D) Amino acid conservation of N-terminal region of Slc25a39 uORF-encoded peptides in different mammalian species (top), conservation of nucleotides around the start site of Slc25a39 uORF in different mammalian species (bottom).
(E) Heatmap plot highlighting transcriptionally regulated genes involved in glutathione synthesis (eIF2αA/A compared with eIF2α+/+ mESCs in LIF). Values are log-transformed z-scored RNA-seq expression level.
(F) UCSC Genome browser views for ATF4 binding events in GCN2+/+ (WT) and GCN2−/− (KO) cells that have been exposed to control (+Leu) or leucine-deficient (−Leu) medium. ATF4 binding profiles in ATF4+/+ (WT) and ATF4−/− (KO) cells treated with tunicamycin (Tm) (GSE35681) are shown below each panel.
(G) The heatmap plot shows fold enrichment for selected ATF4 ChIP-Seq target genes associated with glutathione synthesis.
(H) Metabolomic analysis of eIF2α+/+ and eIF2αA/A mESCs under LIF conditions. Significantly up- and downregulated metabolites (p < 0.05 and −0.5 > log2 FC > 0.5) are highlighted in red and blue, respectively.
(I) Schematic summary of genes that are translationally or transcriptionally regulated by ISR and are involved in glutathione metabolism. CTH, cystathionine gamma-lyase; CHAC1, ChaC glutathione-specific gamma-glutamylcyclotransferase 1; GPX, glutathione peroxidase; GSS, glutathione synthetase; GSSG, glutathione disulfide; GSH, glutathione; Glu, glutamate; Gln, glutamine.
DISCUSSION
Here, we demonstrate that pre-implantation embryos display a spatiotemporal regulation of p-eIF2α in the epiblast layer of E4.5 mouse embryos. We show that, in the absence of p-eIF2α (eIF2αA/A), mESC transition from the naive state of pluripotency is accelerated concomitantly with translational regulation of a subset of mRNAs involved in pluripotency, chromatin organization, and metabolism.
Our data show that mRNAs with high GC content are resistant to translation inhibition by p-eIF2α. The enrichment of mRNAs with short CDS and 3′ UTR among the p-eIF2α-resistant mRNAs agrees with Lodish’s model that a reduced general translation favors the translation of mRNAs with faster translation initiation rates.42 The higher TE of mRNAs with short CDS under translation inhibition is also supported by an inverse relationship between CDS length and TE observed across multiple species.43,70
We documented several p-eIF2α-resistant mRNAs that harbor asingleuORFat their 5′ UTRs. The regulatory role of a single inhibitory uORF in the translation of p-eIF2α-resistant mRNAs such as CHOP was described.71 eIF2α phosphorylation promotes the bypass of some uORFs with a suboptimal start site.71 Here, we provide results that p-eIF2α-dependent regulation of eIF1 expression via ATF4 could contribute to this bypass.
The translational coordination of mRNAs comprising the cohesin and condensin complexes is cell-cycle dependent.72 Importantly, mESCs are highly sensitive to changes in cohesin and condensin levels relative to somatic cells.73-75 Our results underscore the importance of p-eIF2α-dependent translation in controlling the expression of the cohesin and condensin complexes and, consequently, chromosome condensation and segregation.
We report a dramatic p-eIF2α-dependent transcriptional reprogramming downstream of translational control. We also demonstrate that p-eIF2α controls multiple components of the glutathione metabolic pathway. The link between the ISR pathway, glutathione metabolism, and antioxidant response is well documented.68 Disruption of glutathione metabolism negatively impacts pluripotency and differentiation of mESCs.76 The data provide a mechanistic model where p-eIF2α coordinates glutathione metabolism by controlling the biosynthesis of multiple components of the pathway, including cellular and mitochondrial transporters.
The activity of a few translation factors has been linked to pluripotency regulation. Abrogation of eIF4G2/DAP5 in mESCs engenders a naive state pluripotency status.4 eIF4G2/DAP5 interacts with eIF2 and promotes leaky scanning through uORFs.77-79 Like p-eIF2α, and consistent with a lower translation rate in the naive state, depletion of eIF4G2/DAP5 results in reduced global translation.77,80
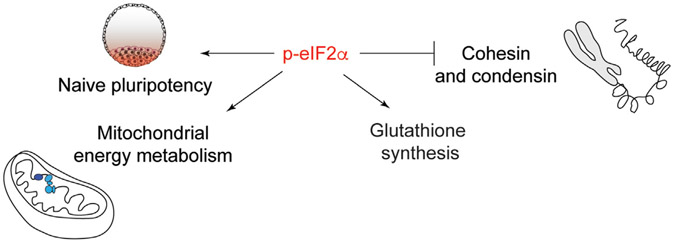
In conclusion, our data demonstrate that p-eIF2α controls translation of genes involved in key cellular processes in pluripotent cells (Figure 7).
Figure 7. Model summarizing the translational regulation of genes involved in key cellular processes by p-eIF2α in pluripotent cells.
Limitations of the study
A clear limitation of our study is the lack of functional analysis of p-eIF2α targets in the maintenance of pluripotency and lineage commitment. It is immediately clear which are the most prominent and actionable targets to study. Nevertheless, we suggest that the best questions to address are the translational coordination of cohesin/condensin by eIF2 vis-a-vis the impact on chromatin organization in mESCs. Also, given the critical role of glutathione in antioxidant defense, how will regulation of glutathione metabolism by p-eIF2α impact redox signaling in mESCs.
It seems counterintuitive that such an important molecular mechanism, which controls the synthesis of key proteins, is not essential for embryogenesis since eIF2αA/A mice survive to birth and do not display any obvious developmental defects. In contrast, sustained increase in p-eIF2α in Ppp1r15a−/−, Ppp1r15b−/− embryos results in early lethality. It is expected that deregulation of the same subset of p-eIF2α-sensitive mRNAs we identified is implicated in the early lethality in Ppp1r15a−/−, Ppp1r15b−/− embryos. Staining of E4.5 embryos demonstrated a transient increase in p-eIF2α in the naive epiblast layer. However, the nature of upstream eIF2 kinase(s) is not known. It is conceivable that the effect of p-eIF2α is important in early development during stress conditions such as nutrient deprivation. For example, GCN2−/− mice appear developmentally normal in non-stressed condition; however, they display severe developmental defect when leucine is absent in the mother’s diet.22
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Nahum Sonenberg (nahum.sonenberg@mcgill.ca).
Materials availability
Materials in this study will be made available on request. Please note that their transfer will be subject to a Materials Transfer Agreement.
Data and code availability
The Ribo-seq data are available in NCBI’s Gene Expression Omnibus (GEO) through GEO Series accession number GEO: GSE248729 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE248729).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Mouse embryonic stem cell derivation and culture
mESCs were derived from eIF2α S51A/+ C57Bl/6J mice as described.91 Briefly, heterozygous mice (eIF2α S51A/+) were crossed, and the resulting embryos were flushed out of the uterus of pregnant mice at 3.5 d.p.c., washed through several drops of M2 medium (Sigma Aldrich, Cat#M7167) and transferred to four-well plates (R&D systems, Cat#PSC001) coated with irradiated mouse embryonic fibroblasts (iMEFs) (one embryo per well). Embryos were maintained in a serum-free ESC media (EmbryoMax DMEM (Millipore, SLM-220-B) supplemented with 10% KnockOut Serum Replacement (Invitrogen, Cat#10828-028), 1X MEM non-essential Amino Acid Solution (Invitrogen, Cat#11140-050), 1 mM Sodium Pyruvate (Invitrogen, Cat#11360-070), 2 mM GlutaMax (Invitrogen, Cat#35050-061), 100 U/mL Penicillin, 100 mg/mL Streptomycin (Invitrogen, Cat#15140-122), 0.11 mM β-mercaptoethanol (Invitrogen, Cat#21985-023), 1 mM PD0325901 (BioVision, Cat#1643-2) and 3 mM CHIR99021 (Stemgent, Cat#04-0004), and 1000 U/mL ESGRO Leukemia Inhibitory Factor (LIF) (EMD Millipore, Cat# ESG1107) at 37°C, 5% CO2 for 48 h. If blastocysts were hatched, the media was changed every other day for 7–10 days, and then cells were disaggregated using 0.05% (w/v) trypsin and passaged the cells 1:4 onto a new iMEF coated plate. Some cells were frozen in freezing media, and the rest were expanded on feeder-free plates coated with 0.1% Gelatin (Millipore, Cat#ES-006-B) containing serum-free ESC media. The genotype of the mESCs was determined using PCR, and the status of p-eIF2α in each line was assessed using WB. For the rest of the experiments, mESCs were cultured in feeder-free plates with serum-free ESC media, passaged every 3–4 days, and disassociated with Accutase (Sigma Aldrich, Cat# A6964) unless specified otherwise.
Human and mouse cell lines culture
Murine N2A (ATCC CCL-131) cells with neuronal and ameboid stem cell-like morphology and HEK293T (ATCC CRL-3216) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin-streptomycin and glutamine at 37°C and 5% CO2.
METHOD DETAILS
Alkaline phosphatase (AP) assay
mESCs were cultured in 6-well plates in the presence or absence of 2i for six days. The Alkaline phosphatase assay was performed using AP Detection Kit (Millipore, Cat# SCR004) according to the manufacturer’s instructions. Briefly, mESCs were fixed with 4% Paraformaldehyde (PFA) in PBS for 2 min. PFA was removed, and cells were washed with 1X Rinse Buffer (TBST: 20 mM Tris-HCl pH 7.4, 0.15M NaCl, 0.05% Tween 20)). The rinse buffer was removed, and stain solution consists of Fast Red Violet (FRV), Naphthol AS-BI phosphate solution and water was added in a 2:1:1 ratio (FRV: Naphthol: water) and incubated for 15 min in the dark. Finally, cells were washed once with 1X Rinse Buffer and then 1X PBS. The plates were scanned for visualization of AP+ cells.
Immunofluorescence and whole mount immunostaining
Immunofluorescence was conducted on cells fixed in 4% paraformaldehyde for 10 min at RT, washed three times with PBS, and blocked for 15 min with 5% FBS in PBS containing 0.1% Triton X-100. After incubation with primary antibodies (Overnight, 4°C in 5% FBS in PBST (PBS containing 0.1% Tween 20), cells were washed three times with PBST and incubated with fluorophore-labeled appropriate secondary antibodies purchased from Jackson ImmunoResearch and counterstained with DAPI (1 μg/mL, 0215754; MP Biomedical). Specimens were analyzed using a Zeiss (Oberkochen, Germany) Axioscope microscope, and images were acquired with a Zeiss Axiocam HRM camera.
For whole-mount immunostaining, E4.5 embryos were flushed or dissected from the uteri of pregnant females. The embryos were fixed for 15 min in 4% PFA in PBS at RT and permeabilized with 0.3% Triton X-100 for 5 min. Blocking was performed with 10% FCS in PBST for at least 30 min or stored at 4°C. Primary antibody incubation was 1/200 dilution in blocking solution at 4°C Overnight. The embryos were washed in blocking solution three times and incubated in secondary antibody solution (1/400) for 2 h at RT. After washing three times, the embryos were individually mounted in a well between two coverslips with DAPI mounting solution.
Atf4 siRNA transfection
siRNA transfections were performed using Lipofectamine 2000 (Invitrogen, CA) according to the protocol provided by the manufacturer. Briefly, 3 × 105 N2A cells were seeded in each well of 6-well plates. After 24 h cells were transfected with 20 nM scramble siRNA (Dharmacon, Cat# D-001810-10-05) or siRNA targeting murine Atf4 (Dharmacon, Cat# L-042737-01-0005) for 5 h. Afterward, the transfection media was removed, rinsed with PBS, and replenished with the regular culture medium. The cells were harvested for RNA and protein extraction 72 h post-transfection.
Atf4 shRNA transduction
The Atf4 shRNA (TRCN0000301646) and control plasmid, obtained from McGill Platform for Cellular Perturbation (MPCP), were co-transfected with psPAX2 (psPAX2 was a gift from Didier Trono (http://n2t.net/addgene:12260; RRID:Addgene_12260) and pMD2.G (pMD2.G was a gift from Didier Trono (http://n2t.net/addgene:12259; RRID:Addgene_12259) Addgene, plasmid 12260)) plasmids in HEK293T cells using Lipofectamine 2000 (Invitrogen, CA). Forty-eight hours post-transfection, supernatant was collected and passed through a 0.45 μM filter. Transduction was performed on 3 × 105 N2A cells, which were seeded a day prior to transduction, in presence of polybrene (6 μg/mL), and antibiotic selection (puromycin, 2 μg/mL) was started two days post-transduction. The knockdown of the Atf4 was confirmed by Western blotting and qPCR.
Polysome isolation and sucrose gradient fractionation
Cells in culture were treated with cycloheximide (CHX) (100 μg/mL) for 5 min at 37°C and then washed two times with cold PBS containing CHX (100 μg/mL). Cells were scraped off the plate using a rubber scraper with residual PBS solution and centrifuged at 4°C for 5 min at 1500 rpm. The pellet was resuspended in 425 mL hypotonic lysis buffer (20 mM Tris-HCl pH 7.4; 150 mM NaCl; 5 mM MgCl2; 1 mM DTT; 100 μg/mL CHX; supplemented with cOmplete; EDTA-free Protease Inhibitor Cocktail). To each cell lysate, 5 μL of RiboLock RNase inhibitor (ThermoFisher Scientific, Cat#EO0382), 5 μL of Turbo DNase (ThermoFisher Scientific, Cat#AM2238), 50 μL of Triton X-100 (10% v/v), and 50 μL of sodium deoxycholate (10% w/v) were added. Cell lysates were vortexed for 5 s and incubated on ice for 10 min followed by centrifugation at 12,000 rpm for 10 min at 4°C. Supernatants were collected and loaded on a pre-chilled 10–50% sucrose gradient generated by Gradient Master (Biocomp Instruments) after the addition of equal volumes of 10% and 50% sucrose solution (in polysome buffer containing 20 mM Tris-HCl pH 7.4; 150 mM NaCl; 5 mM MgCl2; 100 μg/mL CHX; supplemented with cOmplete, EDTA-free Protease Inhibitor Cocktail) in a centrifuge tube (Beckman Coulter). The gradient was centrifuged at 36,000 rpm (SW40 rotor, Beckman Coulter) for 2 h at 4°C. Fractions were collected (14 fractions of 35 s each) using a collector (Brandel) and UV optical unit (Brandel) and stored at −80°C.
RNA extraction from sucrose gradient fractions
Sucrose gradient fractions were subjected to RNA extraction by adding 800 μL of TRIzol (Invitrogen, Cat#15596018) to each fraction. The fractions were then mixed by vigorous shaking and incubated at room temperature (RT) for 5 min 200 μL of chloroform was added to each extract, and mixed before incubating for 2 min at RT. The extracts were then centrifuged at 12,000 rpm for 15 min at 4°C. The colorless upper layer was transferred to a pre-chilled tube containing 90 μL of sodium acetate (3 M pH 5.5) and 2 μL of Glycoblue Coprecipitant (Invitrogen, Cat#AM9516). 500 μL of isopropanol was added to the aqueous phase, and thoroughly mixed immediately by inverting the tube. The RNA mixture was stored at −80°C overnight to maximize precipitation. The frozen RNA solution was centrifuged at 20,000 × g for 35 min at 4°C. The precipitated RNA pellet was washed with ice-cold ethanol (80%) and left to dry at RT. The dried pellet was dissolved in RNase-free water, and the concentration was measured by Nanodrop Spectrophotometer (ThermoFisher Scientific) and adjusted to a final concentration of 250 ng/μL.
cDNA synthesis and quantitative PCR analysis of polysome fractions
RNA extracted from polysome fractions were pooled to generate five pooled fractions (A to E). Reverse transcription was performed using a SuperScript III Reverse-Transcriptase Kit (Invitrogen, Cat#18080044) and Oligo(dT)12-18 (Invitrogen, Cat#18418012) according to the manufacturer’s instructions. Quantitative PCR was performed using SYBR Green (Invitrogen, Cat#4309155) and CFX Connect Real-Time PCR Detection System (Bio-Rad) equipment. Each reaction contained 50 ng of cDNA and 4 fmol of each primer. All experiments were performed in triplicates. A list of primers is provided in Table S5.
Cellular protein extraction
Cells were lysed with RIPA buffer (50 mM Tris-HCl pH 7.4; 1% NP-40; 0.1% SDS; 150 mM NaCl; 2 mM EDTA pH 8) supplemented with protease and phosphatase inhibitors. The lysate was incubated at 4°C with rotation for 30 min and was centrifuged at 15,000 rpm for 15 min at 4°C. Supernatant was collected for protein measurement, followed by Western blotting.
SDS-PAGE and immunoblotting
Proteins were denatured by adding 5X loading buffer (250 mM Tris-HCl pH 6.8; 8% w/v SDS; 0.2% w/v bromophenol blue; 40% v/v glycerol; 20% v/v β-mercaptoethanol) and after boiling subjected to polyacrylamide gel electrophoresis followed by transferring onto a nitrocellulose membrane. Membranes were blocked with 5% skim milk solution (in 0.1% TBST). Western blotting was carried out using the following antibodies and those listed in the key resources table: Antibodies against UTF1, SMC2, SMC4, and eIF2α obtained from Cell Signaling Technologies. Antibodies against KLF2, phospho-eIF2α S51, and β-actin purchased from Abcam Inc. Antibodies against eIF1, ATF4, and OCT3/4 obtained from Santa Cruz Biotechnology and antibodies against NANOG and NCAPG from Bethyl.
Ribosome profiling
Cells were washed twice with ice-cooled PBS containing 100 μg/mL CHX. Residual PBS was aspirated thoroughly, 250 μL of 2X lysis buffer (40 mM Tris-HCl pH 7.4, 300 mM NaCl, 10 mM MgCl2, 200 μg/mL CHX, 2 mM DTT, supplemented with cOmplete Protease Inhibitor Cocktail) was added to the cells. In-dish lysis was performed by scraping the cells with the lysis buffer on ice and transferring the lysates into microfuge tubes. 5 μL of RiboLock RNase inhibitor (ThermoFisher Scientific, Cat#EO0382) and 5 μL of Turbo DNase (ThermoFisher Scientific, Cat#AM2238) were added to the tube. Lysates were vortexed for 5 s, and incubated on ice for 10 min, followed by centrifugation at 16000 × g for 10 min at 4°C. Supernatants were transferred to a new microfuge tube, and OD was measured at A260/280 for each sample by Nanodrop before storing at −80°C. Before nuclear digestion, a portion of each lysate was saved for RNA-seq. Ribo-seq samples were digested with MNase for 45 min at RT. Digestion conditions with MNase were optimized to yield the highest recovery of monosomes after digestion without severe damage to the integrity of ribosomes, as previously described.92 In our optimized condition, 14U MNase per A260 Unit and 6U RNAse I (Epicenter) per A260 Unit were used for 45 min digestion at RT. RNAse I and MNase were inactivated by adding SUPERase In RNase inhibitor and CaCl2, respectively, into the corresponding lysate and incubating for 5 min. Digested polysomes were loaded on a 10–50% sucrose density gradient and centrifuged at 36,000 rpm for 3 h at 4°C using an SW40Ti rotor (Beckman Coulter). Gradients were fractionated as described above, and UV absorbance was monitored. Fractions corresponding to monosome peak fractions were collected, pooled, and RNAs were extracted using Trizol (Thermo Fischer Scientific). For RNA-seq, total RNA was extracted using Trizol from the rest of the lysate.
For Ribo-seq library preparation, the most recently published ribosome profiling technique was used32 with several modifications described below. Briefly, purified RNA samples were separated electrochemically on 15% TBU-Urea gel, and fragments in the range of 25 nt and 40 nt were selected and excised from the gel. RNA samples were recovered from the gel by adding 400 μL RNA gel extraction buffer (300 mM NaOAc pH 5.5, 1 mM EDTA, 0.25% v/v SDS). Next, the mixture was frozen on dry ice for 30 min, followed by gentle rotation at RT overnight. The footprint fragments were purified by isopropanol precipitation. Depletion of rRNA was performed using riboPOOLs according to the manufacturer’s instructions (siTOOLs Biotech, Germany). rRNA-depleted RNA samples were dephosphorylated with T4 Polynucleotides Kinase (T4 PNK, NEB) for 1 h at 37°C in a reaction containing the final concentration of 1X T4 PNK Buffer, 10 U SUPERase*In and 5U T4 PNK (10 U/μL). The ligation reaction was immediately performed by adding the ligation reaction mixture to the samples (17.5% w/v PEG-8000, 1XT4 RNA ligase buffer, 100 U T4 Rnl2(tr) K227Q (200 U/μL) and 1 μM pre-adenylated linkers). The pre-adenylated linkers contain a 5-nt Unique Molecular Identified (UMI) and 5-nt barcode to distinguish between samples and PCR duplicates during library preparation. The ligated samples were separated on a 10% TBE-Urea gel, and the excised gels were then purified using RNA extraction buffer and isopropanol, as described above. The purified ligated RNA samples were reverse-transcribed, starting by adding 2μL reverse transcription primer (containing 2 UMI and an 18-atom hexa-ethyleneglycol spacer to block elongation by DNA polymerases) to the 10 μL ligated RNA samples and incubated at 65°C for 5 min. Then, the reverse transcription reaction mixture (5mM DTT, 20 U SUPERase*In, 20 U SuperScript III, 1X First-Strand Buffer, 0.5 mM dNTP) was added to the sample for 30 min at 55°C followed by adding 2.2 μL 1M NaOH to the reaction and incubating at 70°C for 20 min. Samples were then purified using a DNA extraction buffer (300 mM NaCl, 10 mM Tris pH 8, 1 mM EDTA). Purified cDNA was circularized using CircLigase I (Lucigen), followed by another round of rRNA depletion using biotin-labeled antisense probes and Dynabeads MyOne Streptavidin C1 as previously described.93 Finally, a qPCR was performed to quantify and optimize the amount of circularized cDNA and the cycle number required for library preparation according to the instruction in the protocol. The final PCR reaction was performed using Phusion polymerase, and the PCR product was purified using a DNA clean & concentrator column according to the manufacturer’s instruction. Purified PCR products were separated on an 8% TBE gel, and the bands corresponding to ~160 bp were excised and extracted using the DNA extraction buffer as described above. The resulting library size and quantity were confirmed using a Bioanalyzer High Sensitivity DNA Analysis (Agilent), and libraries were then sequenced using an Illumina NovaSeq 6000 System.
The same procedure was performed for RNA-seq library preparation except for the following changes. rRNA depletion was performed on total RNA using NEBNext rRNA Depletion Kit (Human/Mouse/Rat) strictly according to the manufacturer’s instruction. rRNA-depleted RNAs were fragmented using NEBNext Magnesium RNA Fragmentation Module for 6 min at 94°C. Fragmented RNAs were processed as for Ribo-seq libraries.
Sequencing data alignment
Sequenced libraries were trimmed using Cutadapt 3.481 by removing the constant adapter (AGATCGGAAGAGCAC) from reads without allowing any mismatches. Any reads shorter than six nt and without the adapter were discarded. Trimmed reads were demultiplexed using the sample barcode anchored to the 5′ end of reads without allowing any mismatch in the barcodes. The multiplexed reads were then processed for alignments to the mouse transcriptome using the following criteria: First, low-quality reads and reads shorter than 25 were discarded, then reads were sequentially aligned to cytoplasmic and mitochondrion rRNA and tRNA using Bowtie 1.3.0,82 allowing three mismatches in the reads (parameters: −v 3 −norc −p 12). The remaining reads were aligned to mouse Gencode version M25 comprehensive transcriptome94 with Bowtie 1.3.0 allowing two mismatches with the following parameters: (−norc −a −p 12 −m 100 −n 2 −seedlen 25). SAM files were converted to BAM files, sorted, indexed, and then PCR duplicates were removed using UMI-Tools 1.0.1.83 The final BAM file was converted to an SQLite file compatible with Trips-Viz (https://trips.ucc.ie/), a platform for the Ribo-seq data analysis.84
Metagene profiles and offset determination
Metagene profiles were created by aligning either the annotated CDS start or CDS stop from all coding principal transcript isoforms (as determined by taking the isoform with the longest CDS) such that the CDS start/CDS stop lies at position 0. For CDS start metagene profiles, this analysis was performed on a per read length basis. Offsets were determined by calculating the distance in nucleotides between position 0 (the CDS start) and the highest peak upstream of position 0, 3 nts were then added to this value to infer the A-site of each read.
Differential expression analysis
Read count matrices for both RNA-seq and Ribo-seq were taken from the “Read Table” built-in function of Trips-Vis, which quantifies the reads uniquely aligning to the CDS region of mRNAs using inferred A-sites of reads. If the inferred A-site of a Ribo-seq read lies between the annotated CDS start and CDS stop of a transcript, it contributes to the CDS count. Genomic coordinates of each read were calculated to determine whether the read maps unambiguously. Differential analysis was performed using the R package deltaTE (ΔTE) version 1.34.0,39 which incorporates an interaction model into the DESeq2 method. For each gene, a p-adjusted value (p-adj) was calculated for the fold changes between two experimental conditions at Ribo-seq, RNA-seq, and TE levels, and p-adj below 0.05 was considered significant for each fold change. These fold changes were then used to divide genes into eight categories described in the text. For scatterplots displaying the fold change between Ribo-seq and RNA-seq in Figure 2B, genes with the p-adj value of “NA” in any fold change (Ribo-seq, RNA-seq, and TE) comparison were excluded from the graph. The values for the PCA plots in Figure 2B were also determined using DESeq2.
uORF analysis
The 5′UTR sequences of every principal transcript isoform (the isoform with the longest CDS) were parsed to find the coordinates of each uORF (AUG > STOP). If an ORF had multiple AUGs it was only counted once. The number of RNAse I generated Ribo-seq reads supporting inferred A-sites with locations between the uORF start and stop codons was counted. The ORF was considered translated if more than 5 Ribo-seq reads were present. In the case of multiple ORFs having more than 5 Ribo-seq reads, the ORF with the highest number of reads was used. Translation efficiency (TE) was calculated for the translated ORFs by taking the RPKM of MNase digested Ribo-seq reads within the ORF and dividing it by the RPKM of RNA-seq reads aligning to the entire transcript.
Gene features
The lengths of 5′UTRs/CDS regions/3′UTRs were from Gencode version M25 GTF annotations. GC% for each region was calculated for each region using an in-house python script. Minimum free energy (MFE) was calculated using the python package Vienna RNA version 2.1.9.85 Sequences of length <40 nucleotides and >50000 nucleotides were discarded. MFE values were normalized for sequence length using the formula 100*(MFE/Sequence length).
For all ribosome coverage plots, CDS and uORF regions are shown by thick black and green lines, respectively. Read numbers are the aggregate of two replicates for each condition from RNase I digested libraries.
ChIP-seq data analysis
ATF4 ChIP-Seq analysis was described previously,59 and associated datasets are available in the Gene Expression Omnibus (GEO) database under the accession number (GSE166590). Briefly, raw reads were trimmed for length (n ≥ 50), quality (phred score ≥ 30), and adapter sequence using Fastx v0.0.13.2 (FASTQ/A short-reads pre-processing tools, http://hannonlab.cshl.edu/fastx_toolkit/). Trimmed reads were then aligned to the mouse reference genome mm10 using BWA v0.7.12.95 Peaks were called using MACS (Model-based Analysis of ChIP-Seq) v2.1.0 software and default parameters (mfold = [5,50]; FDR cutoff = 0.05, −nomodel)96 using sequenced libraries of either WT or GCN2 KO input DNA as control. Peak annotations and bed files were generated using HOMER (Hypergeometric Optimization of Motif EnRichment) v4.7.97 For peak intersections, peak list intersections were performed using the R package GenomicRanges v1.26.4.98 Binding peaks were considered overlapping if their peak summits were within 300 bp or less apart. ChIP-seq tracks were visualized using the UCSC Genome Browser (https://genome.ucsc.edu/).
Preparation of samples for mass spectrometry analysis
Cultured cells were rinsed with phosphate buffer saline (PBS) and scraped into lysis buffer containing 8 M urea, 20 mM HEPES-KOH pH 8.5, with 2X phosphatase inhibitors (Cell Signaling Technology #5870). Samples were probe tip sonicated, reduced with 5 mM DTT for 50 min at 55°C, alkylated for 30 min with 10 mM iodoacetamide, and quenched with 5 mM DTT. Samples were diluted to 2M urea with digestion dilution buffer (20 mM HEPES-KOH pH 8.5 containing 1 mM CaCl2) and digested at 37°C with 20 μg of Lysyl Endopeptidase (Wako-Chem). Samples were diluted to 1 M urea and digested for 5 h with 20 mg of trypsin (Pierce). Following digestion, peptides were acidified with trifluoroacetic acid (TFA), centrifuged at 500 × g for 20 min and purified over SepPak C18 columns. Following elution, peptides were quantified with a MicroBCA assay (Thermo Fisher Scientific, San Jose, CA).
Total protein sample preparation
20 μg of peptides from each sample were labeled with isobaric tandem-mass-tag (TMT) 16 plex reagents (Thermo Fisher Scientific, San Jose, CA) in 20 mM HEPES-KOH pH 8.5 with 30% acetonitrile (v/v) with 50 μg of TMT reagent. The reaction was quenched for 15 min by adding hydroxylamine to a final concentration of 0.3% (v/v). Samples were combined, dried, purified over SepPak C18 columns, and dried again. Samples were then resuspended in 40 μL of basic reverse phase (bRP) buffer A (10 mM NH4HCO2 pH 10, 5% acetonitrile (CAN)) and separated on a Zorbax Extended C18 column (2.1 × 150 mm, 3.5 mm, no. 763750-902, Agilent) using a gradient of 10–40% bRP buffer B (10 mM NH4HCO2, pH 10, 90% ACN). 96 fractions were collected before concatenation 24 fractions. Each fraction was dried and desalted over a C18 STAGE-Tip prior to analysis by mass spectrometry.
IMAC phosphopeptide sample preparation
100 μg of peptides from each sample were labeled with isobaric tandem-mass-tag (TMT) 16 plex reagents (Thermo Fisher Scientific, San Jose, CA) in 20 mM HEPES pH 8.5 with 30% acetonitrile (v/v) with 200 μg of TMT reagent. The reaction was quenched for 15 min by adding hydroxylamine to a final concentration of 0.3% (v/v). Samples were combined, dried, purified over SepPak C18 columns, and dried again. High-Select Fe-NTA Phosphopeptide Enrichment Kits from Thermo were used to enrich phosphopeptides from TMT-labeled peptides. Following the elution from the IMAC column, enriched samples were dried, purified over SepPak C18 columns, and bRP fractionated as described above instead with a gradient of 5–40% bRP buffer B. The 12 concatenated fractions were desalted over a C18 STAGE-Tip prior to analysis by mass spectrometry.
LC-MS analysis of total protein and IMAC fractions
Samples were analyzed on an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific, San Jose, CA) coupled with a Proxeon EASY-nLC 1200 liquid chromatography (LC) pump (Thermo Fisher Scientific, San Jose, CA). Peptides were separated on a 100 μm inner diameter microcapillary column packed with ~40 cm of Accucore150 resin (2.6 μm, 150 Å, ThermoFisher Scientific, San Jose, CA). For each analysis, 1 mg was loaded onto the column. Peptides were separated using 2.5 h gradient of 6–30% ACN in 0.125% formic acid with a flow rate of 550 nL/min. Each analysis used an SPS-MS3-based TMT method,28,99,100 which has been shown to reduce ion interference compared to MS2 quantification.101 The scan sequence began with an MS1 spectrum (Orbitrap analysis, resolution 120,000; 350–1400 m/z, automatic gain control (AGC) target 4.0 × 105, maximum injection time 50 ms). Precursors for MS2/MS3 analysis were selected using a Top10 method. MS2 analysis consisted of collision-induced dissociation (quadrupole ion trap; AGC 2.0 × 104; normalized collision energy (NCE) 35; maximum injection time 120 ms). Following the acquisition of each MS2 spectrum, we collected an MS3 spectrum, a method in which multiple MS2 fragment ions are captured in the MS3 precursor population using isolation waveforms with multiple frequency notches.100 MS3 precursors were fragmented by HCD and analyzed using the Orbitrap (NCE 65, AGC 3.5 × 105, maximum injection time 150 ms, isolation window 1.2 Th, the resolution was 50,000 at 200 Th).
Total protein data processing and analysis
Mass Spectra were processed using a Comet-base software pipeline.86,87 Data were searched with a fully tryptic database containing human Swissprot consensus entries plus isoforms, allowing for a static modification of lysine and N-termini with TMT (304.2071 Da) and carbamidomethylation (57.0215 Da) of cysteine, along with variable oxidation (15.9949 Da) of methionine. Searches were performed using a 20 ppm precursor ion tolerance, and the product ion tolerance was set to 1.0 Th. Peptide-spectrum matches (PSMs) were adjusted to a 1% false discovery rate (FDR) using previously described linear discriminant analysis.88,89 Filtered PSMs were collapsed to a final protein-level FDR of <1%. Protein assembly was guided by principles of parsimony to produce the smallest set of proteins necessary to account for all observed peptides.87 For TMT-based reporter ion quantitation, we extracted the summed signal-to-noise (S/N) ratio for each TMT channel. We found the closest matching centroid to the expected mass of the TMT reporter ion. MS3 spectra with TMT reporter ion summed S/N ratios less than 250 were excluded from quantitation.102 The list of channels, proteins, and their S/N ratios for downstream analysis are provided in Table S3.
Phosphorylation data processing and analysis
For searching phosphorylation data, a variable modification for phosphorylation (79.9663 Da) was allowed on serine, threonine, and tyrosine. Searches were performed using a 20-ppm precursor ion tolerance, and the product ion tolerance was set to 0.02 Th. Linear discriminant analysis (LDA) was performed to set a PSM FDR of <1%. Filtered PSMs were collapsed to a final protein-level FDR of <1%. Phosphorylation sites were evaluated using the AScore method, and sites with an AScore >13 were considered confidently localized.90 PSMs with TMT reporter ion summed S/N ratios less than 250 were excluded from quantitation. The list of channels, phosphoproteins, and their S/N ratios for downstream analysis are provided in Table S3.
Metabolome analysis
Cells are washed twice with ice-cold normal saline solution. 0.8 mL 80% HPLC-grade Methanol was added to the cells and scraped onto an eppendorf tube. Cells were subjected to two rounds of 10min sonication, 30s on 30s off using automatic sonicator followed by centrifugation at 12000 rpm for 10 min at 4°C. Supernatants collected from mESCs were transferred to new pre-chilled tubes containing 1 μL of 800 ng/μL 2H27-Myristic in pyridine. Samples were then dried by vacuum centrifugation operating at a sample temperature of −4°C.
After drying, samples were subjected to a two-step derivatization for GC/MS analysis. First, the samples were resuspended in 30 μL of 10 mg/mL Methoxyamine: HCl in anhydrous pyridine (MOX), followed by sonication and vortex for 15 s three times. After centrifugation for 3 min at 15,000 rpm (RT), samples were incubated for 30 min at RT. The samples were then centrifuged for 2 min at 15000 rpm and transferred to 2 mL GC/MS sample vials containing 250 μL glass inserts with 70 μL of N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide (MTBSTFA). Following GC/MS scan and single ion monitoring (SIM) data acquisition, samples were diluted using 30% MOX and 70% MTBSTFA 1:25 and data were acquired in scan mode.
An Agilent 5975C GC-MS equipped with a DB-5MS + DG (30 m × 250 μm x 0.25 μm) capillary column (Agilent J&W, Santa Clara, CA, USA) was used for all GC-MS experiments, and data collected by electron impact set at 70 eV both in scan (50–1000 m/z) and single ion monitoring modes. A total of 1 μL of the derivatized sample was injected per run in spitless mode with an inlet temperature set to 280°C, using helium as a carrier gas with a flow rate such that (2H27− myristic acid 18 min. The quadrupole was set at 150°C and the GC/MS interface at 285°C. The oven program for all metabolite analyses started at 60°C held for 1 min, then increasing at a rate of 10°C/min until 320°C. Bake-out was at 320°C for 10 min. Sample data were acquired in scan mode (50–1000 m/z) or SIM with a 5 ms dwell time where the M-57 [M+.−C4H9.]+ fragment was used for relative quantitation (area under the curve) in both modes of data acquisition. Data were analyzed using Mass Hunter Quant (Agilent Technologies). The reported spectra and retention times of all metabolites were confirmed by methoxylamine and tert-butyldimethylsilylated authentic standards. The area under the curve was normalized to cell numbers.
QUANTIFICATION AND STATISTICAL ANALYSIS
Sample sizes and statistical tests used in the paper are described in the figure legends and were performed with Prism 10 (GraphPad), RStudio and DESeq2 software. Differences were considered statistically significant at a p value < 0.05. For multiple comparison analyses, appropriate post hoc tests were used to access statistical difference between specific groups and described in the figure legends. Unless otherwise stated, values shown are mean ± SEM and statistical treatments are One-way, two-way ANOVA followed by appropriate post hoc comparisons or unpaired t test with Welch correction as indicated in the figure legends.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal UTF1 | Cell Signaling Technology | Cat#3909; RRID:AB_2288387 |
| Mouse monoclonal KLF4 | Santa Cruz Biotechnology | Cat#166238; RRID:AB_2130234 |
| Rabbit monoclonal eIF2α (Clone (D7D3) XP®) | Cell Signaling Technology | Cat#5324; RRID:AB_10692650 |
| Rabbit monoclonal phospho-eIF2α (Ser51) | Abcam Inc. | Cat#ab32157; RRID:AB_732117 |
| Mouse monoclonal ATF4 (Clone B-3) | Santa Cruz Biotechnology | Cat#sc-390063; RRID:AB_2810998 |
| Mouse monoclonal eIF1 (Clone B2) | Santa Cruz Biotechnology | Cat#390122 |
| Mouse monoclonal β-actin | Abcam | Cat#ab179467; RRID:AB_2737344 |
| Mouse monoclonal OCT3/4 | Santa Cruz Biotechnology | Cat#sc-101534; RRID:AB_2237389 |
| Rabbit polyclonal NANOG | Bethyl | Cat#A300-397A; RRID:AB_386108 |
| Mouse monoclonal NCAPG | Bethyl | Cat#A300-602A; RRID:AB_2149992 |
| Rabbit monoclonal SMC4 (Clone D14E2) | Cell Signaling Technology | Cat#5547; RRID:AB_10698892 |
| Rabbit monoclonal SMC2 (Clone D91E3) | Cell Signaling Technology | Cat#5394; RRID:AB_10693943 |
| Goat polyclonal SOX17 | R&D Systems | Cat#AF1924; RRID:AB_355060 |
| Goat polyclonal GATA6 | R&D Systems | Cat#AF1700; RRID:AB_2108901 |
| Chemicals, peptides, and recombinant proteins | ||
| PD0325901 | BioVision | Cat#1643-2 |
| CHIR99021 | Stemgent | Cat#04-0004 |
| ESGRO Leukemia Inhibitory Factor (LIF) | Millipore | Cat#ESG1107 |
| Lysyl Endopeptidase Mass Spectrometry Grade (Lys-C) | Wako Chemicals USA | Cat#NC9223464 |
| Pierce™ Trypsin Protease, MS Grade | ThermoFisher Scientific | Cat#90058 |
| DTT (dithiothreitol) | ThermoFisher Scientific | Cat#R0861 |
| Iodoacetamide | Sigma Aldrich | Cat#A3221 |
| Pierce™ Trifluoroacetic Acid (TFA), LC-MS Grade | ThermoFisher Scientific | Cat#85183 |
| Phosphatase Inhibitor Cocktail (100X) #5870 | Cell Signaling Technology | Cat#5870 |
| DAPI | MP Biomedical | Cat#157574 |
| Lipofectamine 2000 | Invitrogen | Cat#11668019 |
| T4 Polynucleotide Kinase | New England BioLabs | Cat#M0201S |
| SUPERase·In™ RNase Inhibitor | Invitrogen | Cat#AM2694 |
| CircLigase ssDNA Ligase | Lucigen | Cat#CL4111K |
| Micrococcal nuclease | Sigma Aldrich | Cat#10107921001 |
| TURBO™ DNase | ThermoFisher Scientific | Cat#AM2238 |
| SYBR™ Green PCR Master Mix | Invitrogen | Cat#4309155 |
| T4 RNA Ligase 2, truncated K227Q | New England BioLabs | Cat#M0351L |
| Critical commercial assays | ||
| Alkaline Phosphatase (AP) Detection Kit | Sigma Aldrich | Cat#SCR004 |
| SuperScript™ III Reverse Transcriptase Kit | Invitrogen | Cat#18080044 |
| NEBNext® rRNA Depletion Kit | New England BioLabs | Cat#E6310L |
| NEBNext® Magnesium RNA Fragmentation Module | New England BioLabs | Cat#E6150S |
| TMTpro™ 16plex Label Reagent Set | ThermoFisher Scientific | Cat#A44520 |
| High Select™ Phosphopeptide Enrichment Kit | ThermoFisher Scientific | Cat#A32992 |
| Deposited data | ||
| mESC Ribo-Seq and RNA-Seq data | This Study | GEO: GSE248729 |
| mESC MS data | This Study | Tables S3 and S4 |
| Experimental models: Cell lines | ||
| Mouse: mESCs (derived from eIF2αS51A/+ mice) | Scheuner et al., 200120 | PMID: 11430820 |
| Human: 293T | ATCC | Cat#CRL-3216 |
| Mouse: N2A | ATCC | Cat#CCL-131 |
| Experimental Models: Organisms/Strains | ||
| Mouse: eIF2α S51A/+: B6; 129-Eif2s1tm1Rjk/J | Scheuner et al., 200120 | PMID: 11430820 |
| Oligonucleotides | ||
| Primers for RT-qPCR and PCR. see Table S5 | This Paper | Table S5 |
| Scramble siRNA | Dharmacon Reagents | Cat#D-001810-10-05 |
| siRNA targeting ATF4 | Dharmacon Reagents | Cat#L-042737-01-0005 |
| Recombinant DNA | ||
| psPAX2 | Didier Trono Lab: Packaging and Envelope Plasmids (unpublished) | Addgene plasmid # 12260 |
| pMD2.G | Didier Trono Lab: Packaging and Envelope Plasmids (unpublished) | Addgene plasmid # 12259 |
| pLKO.1-puro Empty Vector Control Plasmid DNA | Sigma-Aldrich | SHC001 |
| PLKO.1-shATF4 Plasmid (TRCN0000301646) | McGill Platform for Cellular Perturbation (MPCP) | N/A |
| Software and algorithms | ||
| Cutadapt 3.4 | Martin, 201181 | http://code.google.com/p/cutadapt/ |
| Bowtie 1.3.0 | Langmead et al., 200982 | http://bowtie.cbcb.umd.edu |
| Mouse Gencode version M25 comprehensive transcriptome | Gencode | https://ftp.ebi.ac.uk/pub/databases/ gencode/Gencode_mouse/release_M25/ |
| UMI-Tools 1.0.1 | Smith et al., 201783 | https://GitHub.com/CGATOxford/UMI-tools |
| Trips-Viz | Kiniry et al., 202184 | https://trips.ucc.ie/ |
| R package deltaTE (DTE) version 1.34.0 | Chothani et al., 201939 | https://github.com/SGDDNB/translational_regulation |
| Python package Vienna RNA version 2.1.9 | Lorenz et al., 201185 | http://www.tbi.univie.ac.at/RNA |
| Comet-base software pipeline | Eng et al., 201386; Huttlin et al., 201087 | https://comet-ms.sourceforge.net/https://phosphomouse.hms.harvard.edu/ |
| Linear Discriminant Analysis | Elias and Gygi, 200788; 201089 | PMID: 17327847, 20013364 |
| AScore | Beausoleil et al., 200690 | http://ascore.med.harvard.edu/ |
| Fastx v0.0.13.2 | Cold Spring Harbor Laboratory | http://hannonlab.cshl.edu/fastx_toolkit/ |
Highlights.
spatiotemporal regulation of p-eIF2α in preimplantation mouse embryo
Identification of p-eIF2α targets in mESCs by ribosome profiling
Proportional translation via p-eIF2α in mESCs (cohesin/condensin complexes)
Regulation of glutathione metabolism by p-eIF2α in mESCs
ACKNOWLEDGMENTS
We thank Annie Sylvestre, Annik Lafrance, and Eva Migon for technical assistance and animal handling. This work was funded by the Foundation grant from Canadian Institutes of Health Research (CIHR) (FND-148423) awarded to N.S., the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust and NIH 1R01HL163806-01 awarded to S.T., the Foundation Grant from the CIHR (FDT-156254), and a Program Project Grant from the Terry Fox Research Institute (TFRI) awarded to V.G., and support from NIH. R01 DK113171, P01 HL160472, and R01 CA198103 to R.J.K. GC/MS analyses were carried out at the Metabolomics Core Facility at the Rosalind and Morris Goodman Cancer Institute (GCI), McGill University, Montreal, QC. The Facility is supported by the TFRI, Quebec Breast Cancer Foundation, Fraser Family Trust, McGill Faculty of Medicine, and the GCI. RNA-seq and Ribo-seq libraries in this paper were sequenced on an Illumina NovaSeq 6000 System at Novogene Inc., Sacramento, CA, USA, and at the Donnelly Sequencing Center, Toronto, Canada.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.113615.
REFERENCES
- 1.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, and Smith A (2008). The ground state of embryonic stem cell self-renewal. Nature 453, 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rossant J. (2008). Stem cells and early lineage development. Cell 132, 527–531. [DOI] [PubMed] [Google Scholar]
- 3.Nichols J, and Smith A (2009). Naive and primed pluripotent states. Cell Stem Cell 4, 487–492. [DOI] [PubMed] [Google Scholar]
- 4.Yamanaka S, Zhang XY, Maeda M, Miura K, Wang S, Farese RV Jr., Iwao H, and Innerarity TL (2000). Essential role of NAT1/p97/DAP5 in embryonic differentiation and the retinoic acid pathway. EMBO J. 19, 5533–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li D, Yang J, Huang X, Zhou H, and Wang J (2022). eIF4A2 targets developmental potency and histone H3.3 transcripts for translational control of stem cell pluripotency. Sci. Adv 8, eabm0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, and Selbach M (2011). Global quantification of mammalian gene expression control. Nature 473, 337–342. [DOI] [PubMed] [Google Scholar]
- 7.Sonenberg N, and Hinnebusch AG (2009). Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinnebusch AG, Ivanov IP, and Sonenberg N (2016). Translational control by 5’-untranslated regions of eukaryotic mRNAs. Science 352, 1413–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merrick WC, and Pavitt GD (2018). Protein Synthesis Initiation in Eukaryotic Cells. Cold Spring Harbor Perspect. Biol 10, a033092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa-Mattioli M, and Walter P (2020). The integrated stress response: From mechanism to disease. Science 368, eaat5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dey M, Trieselmann B, Locke EG, Lu J, Cao C, Dar AC, Krishnamoorthy T, Dong J, Sicheri F, and Dever TE (2005). PKR and GCN2 kinases and guanine nucleotide exchange factor eukaryotic translation initiation factor 2B (eIF2B) recognize overlapping surfaces on eIF2αlpha. Mol. Cell Biol 25, 3063–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jousse C, Oyadomari S, Novoa I, Lu P, Zhang Y, Harding HP, and Ron D (2003). Inhibition of a constitutive translation initiation factor 2alpha phosphatase, CReP, promotes survival of stressed cells. J. Cell Biol 163, 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novoa I, Zeng H, Harding HP, and Ron D (2001). Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2αlpha. J. Cell Biol 153, 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andreev DE, O’Connor PBF, Fahey C, Kenny EM, Terenin IM, Dmitriev SE, Cormican P, Morris DW, Shatsky IN, and Baranov PV (2015). Translation of 5’ leaders is pervasive in genes resistant to eIF2 repression. Elife 4, e03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delépine M, Nicolino M, Barrett T, Golamaully M, Lathrop GM, and Julier C (2000). EIF2AK3, encoding translation initiation factor 2-alpha kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat. Genet 25, 406–409. [DOI] [PubMed] [Google Scholar]
- 16.Borck G, Shin BS, Stiller B, Mimouni-Bloch A, Thiele H, Kim JR, Thakur M, Skinner C, Aschenbach L, Smirin-Yosef P, et al. (2012). eIF2gamma mutation that disrupts eIF2 complex integrity links intellectual disability to impaired translation initiation. Mol. Cell 48, 641–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu PJ, Khatiwada S, Cui Y, Reineke LC, Dooling SW, Kim JJ, Li W, Walter P, and Costa-Mattioli M (2019). Activation of the ISR mediates the behavioral and neurophysiological abnormalities in Down syndrome. Science 366, 843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leegwater PA, Vermeulen G, Könst AA, Naidu S, Mulders J, Visser A, Kersbergen P, Mobach D, Fonds D, van Berkel CG, et al. (2001). Subunits of the translation initiation factor eIF2B are mutant in leukoencephalopathy with vanishing white matter. Nat. Genet 29, 383–388. [DOI] [PubMed] [Google Scholar]
- 19.Tahmasebi S, Khoutorsky A, Mathews MB, and Sonenberg N (2018). Translation deregulation in human disease. Nat. Rev. Mol. Cell Biol 19, 791–807. [DOI] [PubMed] [Google Scholar]
- 20.Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, and Kaufman RJ (2001). Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7, 1165–1176. [DOI] [PubMed] [Google Scholar]
- 21.Harding HP, Zhang Y, Scheuner D, Chen JJ, Kaufman RJ, and Ron D (2009). Ppp1r15 gene knockout reveals an essential role for translation initiation factor 2 alpha (eIF2αlpha) dephosphorylation in mammalian development. Proc. Natl. Acad. Sci. USAUSA 106, 1832–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang P, McGrath BC, Reinert J, Olsen DS, Lei L, Gill S, Wek SA, Vattem KM, Wek RC, Kimball SR, et al. (2002). The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol. Cell Biol 22, 6681–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friend K, Brooks HA, Propson NE, Thomson JA, and Kimble J (2015). Embryonic Stem Cell Growth Factors Regulate eIF2αlpha Phosphorylation. PLoS One 10, e0139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kehoe DE, Lock LT, Parikh A, and Tzanakakis ES (2008). Propagation of embryonic stem cells in stirred suspension without serum. Biotechnol. Prog. 24, 1342–1352. [DOI] [PubMed] [Google Scholar]
- 25.Martin Gonzalez J, Morgani SM, Bone RA, Bonderup K, Abelchian S, Brakebusch C, and Brickman JM (2016). Embryonic Stem Cell Culture Conditions Support Distinct States Associated with Different Developmental Stages and Potency. Stem Cell Rep. 7, 177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, Bobrovnikova-Marjon E, Diehl JA, Ron D, and Koumenis C (2010). The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 29, 2082–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalkan T, Olova N, Roode M, Mulas C, Lee HJ, Nett I, Marks H, Walker R, Stunnenberg HG, Lilley KS, et al. (2017). Tracking the embryonic stem cell transition from ground state pluripotency. Development 144, 1221–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zismanov V, Chichkov V, Colangelo V, Jamet S, Wang S, Syme A, Koromilas AE, and Crist C (2016). Phosphorylation of eIF2αlpha Is a Translational Control Mechanism Regulating Muscle Stem Cell Quiescence and Self-Renewal. Cell Stem Cell 18, 79–90. [DOI] [PubMed] [Google Scholar]
- 29.Das I, Krzyzosiak A, Schneider K, Wrabetz L, D’Antonio M, Barry N, Sigurdardottir A, and Bertolotti A (2015). Preventing proteostasis diseases by selective inhibition of a phosphatase regulatory subunit. Science 348, 239–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krzyzosiak A, Sigurdardottir A, Luh L, Carrara M, Das I, Schneider K, and Bertolotti A (2018). Target-Based Discovery of an Inhibitor of the Regulatory Phosphatase PPP1R15B. Cell 174, 1216–1228.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jewer M, Lee L, Leibovitch M, Zhang G, Liu J, Findlay SD, Vincent KM, Tandoc K, Dieters-Castator D, Quail DF, et al. (2020). Translational control of breast cancer plasticity. Nat. Commun 11, 2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGlincy NJ, and Ingolia NT (2017). Transcriptome-wide measurement of translation by ribosome profiling. Methods 126, 112–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ingolia NT, Ghaemmaghami S, Newman JRS, and Weissman JS (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ingolia NT, Lareau LF, and Weissman JS (2011). Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerashchenko MV, and Gladyshev VN (2017). Ribonuclease selection for ribosome profiling. Nucleic Acids Res. 45, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miettinen TP, and Björklund M (2015). Modified ribosome profiling reveals high abundance of ribosome protected mRNA fragments derived from 3’ untranslated regions. Nucleic Acids Res. 43, 1019–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calviello L, Mukherjee N, Wyler E, Zauber H, Hirsekorn A, Selbach M, Landthaler M, Obermayer B, and Ohler U (2016). Detecting actively translated open reading frames in ribosome profiling data. Nat. Methods 13, 165–170. [DOI] [PubMed] [Google Scholar]
- 38.Dunn JG, Foo CK, Belletier NG, Gavis ER, and Weissman JS (2013). Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. Elife 2, e01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chothani S, Adami E, Ouyang JF, Viswanathan S, Hubner N, Cook SA, Schafer S, and Rackham OJL (2019). deltaTE: Detection of Translationally Regulated Genes by Integrative Analysis of Ribo-seq and RNA-seq Data. Curr. Protoc. Mol. Biol 129, e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang T, Cui Y, Jin J, Guo J, Wang G, Yin X, He QY, and Zhang G (2013). Translating mRNAs strongly correlate to proteins in a multivariate manner and their translation ratios are phenotype specific. Nucleic Acids Res. 41, 4743–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lodish HF (1974). Model for the regulation of mRNA translation applied to haemoglobin synthesis. Nature 251, 385–388. [DOI] [PubMed] [Google Scholar]
- 43.Thompson MK, Rojas-Duran MF, Gangaramani P, and Gilbert WV (2016). The ribosomal protein Asc1/RACK1 is required for efficient translation of short mRNAs. Elife 5, e11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaikwad S, Ghobakhlou F, Young DJ, Visweswaraiah J, Zhang H, and Hinnebusch AG (2021). Reprogramming of translation in yeast cells impaired for ribosome recycling favors short, efficiently translated mRNAs. Elife 10, e64283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, and Parker R (2017). The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Mol. Cell 68, 808–820.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Courel M, Clément Y, Bossevain C, Foretek D, Vidal Cruchez O, Yi Z, Bénard M, Benassy MN, Kress M, Vindry C, et al. (2019). GC content shapes mRNA storage and decay in human cells. Elife 8, e49708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torrence ME, MacArthur MR, Hosios AM, Valvezan AJ, Asara JM, Mitchell JR, and Manning BD (2021). The mTORC1-mediated activation of ATF4 promotes protein and glutathione synthesis downstream of growth signals. Elife 10, e63326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Selvarajah B, Azuelos I, Platé M, Guillotin D, Forty EJ, Contento G, Woodcock HV, Redding M, Taylor A, Brunori G, et al. (2019). mTORC1 amplifies the ATF4-dependent de novo serine-glycine pathway to supply glycine during TGF-beta1-induced collagen biosynthesis. Sci. Signal 12, eaav3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J, Yuan CL, Krokowski D, Wang S, Hatzoglou M, et al. (2013). ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol 15, 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li GW, Burkhardt D, Gross C, and Weissman JS (2014). Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiss CS, Ochs MM, Hagenmueller M, Streit MR, Malekar P, Riffel JH, Buss SJ, Weiss KH, Sadoshima J, Katus HA, and Hardt SE (2013). DYRK2 negatively regulates cardiomyocyte growth by mediating repressor function of GSK-3beta on eIF2Bepsilon. PLoS One 8, e70848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Proud CG (2006). Regulation of protein synthesis by insulin. Biochem. Soc. Trans 34, 213–216. [DOI] [PubMed] [Google Scholar]
- 53.Chi B, Wang Q, Wu G, Tan M, Wang L, Shi M, Chang X, and Cheng H (2013). Aly and THO are required for assembly of the human TREX complex and association of TREX components with the spliced mRNA. Nucleic Acids Res. 41, 1294–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vattem KM, and Wek RC (2004). Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. USAUSA 101, 11269–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liefke R, Karwacki-Neisius V, and Shi Y (2016). EPOP Interacts with Elongin BC and USP7 to Modulate the Chromatin Landscape. Mol. Cell 64, 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beringer M, Pisano P, Di Carlo V, Blanco E, Chammas P, Vizán P, Gutiérrez A, Aranda S, Payer B, Wierer M, and Di Croce L (2016). EPOP Functionally Links Elongin and Polycomb in Pluripotent Stem Cells. Mol. Cell 64, 645–658. [DOI] [PubMed] [Google Scholar]
- 57.Ivanov IP, Loughran G, Sachs MS, and Atkins JF (2010). Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1). Proc. Natl. Acad. Sci. USAUSA 107, 18056–18060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loughran G, Sachs MS, Atkins JF, and Ivanov IP (2012). Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res. 40, 2898–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Farooq Z, Kusuma F, Burke P, Dufour CR, Lee D, Tabatabaei N, Toboz P, Radovani E, Greenblatt JF, Rehman J, et al. (2022). The amino acid sensor GCN2 suppresses terminal oligopyrimidine (TOP) mRNA translation via La-related protein 1 (LARP1). J. Biol. Chem 298, 102277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang P, Humphrey SJ, Cinghu S, Pathania R, Oldfield AJ, Kumar D, Perera D, Yang JYH, James DE, Mann M, and Jothi R (2019). Multi-omic Profiling Reveals Dynamics of the Phased Progression of Pluripotency. Cell Syst. 8, 427–445.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lim S, Bhinge A, Bragado Alonso S, Aksoy I, Aprea J, Cheok CF, Calegari F, Stanton LW, and Kaldis P (2017). Cyclin-Dependent Kinase-Dependent Phosphorylation of Sox2 at Serine 39 Regulates Neurogenesis. Mol. Cell Biol 37, e00201–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meister A, and Anderson ME (1983). Glutathione. Annu. Rev. Biochem 52, 711–760. [DOI] [PubMed] [Google Scholar]
- 63.Wang Y, Yen FS, Zhu XG, Timson RC, Weber R, Xing C, Liu Y, Allwein B, Luo H, Yeh HW, et al. (2021). SLC25A39 is necessary for mitochondrial glutathione import in mammalian cells. Nature 599, 136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi X, Reinstadler B, Shah H, To TL, Byrne K, Summer L, Calvo SE, Goldberger O, Doench JG, Mootha VK, and Shen H (2022). Combinatorial GxGxE CRISPR screen identifies SLC25A39 in mitochondrial glutathione transport linking iron homeostasis to OXPHOS. Nat. Commun 13, 2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Young SK, Willy JA, Wu C, Sachs MS, and Wek RC (2015). Ribosome Reinitiation Directs Gene-specific Translation and Regulates the Integrated Stress Response. J. Biol. Chem 290, 28257–28271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ruprecht JJ, and Kunji ERS (2020). The SLC25 Mitochondrial Carrier Family: Structure and Mechanism. Trends Biochem. Sci 45, 244–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lindhurst MJ, Fiermonte G, Song S, Struys E, De Leonardis F, Schwartzberg PL, Chen A, Castegna A, Verhoeven N, Mathews CK, et al. (2006). Knockout of Slc25a19 causes mitochondrial thiamine pyrophosphate depletion, embryonic lethality, CNS malformations, and anemia. Proc. Natl. Acad. Sci. USAUSA 103, 15927–15932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. (2003). An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633. [DOI] [PubMed] [Google Scholar]
- 69.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, and Ron D (2000). Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108. [DOI] [PubMed] [Google Scholar]
- 70.Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, and Herschlag D (2003). Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USAUSA 100, 3889–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Palam LR, Baird TD, and Wek RC (2011). Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J. Biol. Chem 286, 10939–10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stumpf CR, Moreno MV, Olshen AB, Taylor BS, and Ruggero D (2013). The translational landscape of the mammalian cell cycle. Mol. Cell 52, 574–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fazzio TG, and Panning B (2010). Condensin complexes regulate mitotic progression and interphase chromatin structure in embryonic stem cells. J. Cell Biol 188, 491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu G, Kim J, Xu Q, Leng Y, Orkin SH, and Elledge SJ (2009). A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 23, 837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. (2010). Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xin Y, Wang Y, Zhong L, Shi B, Liang H, and Han J (2019). Slc25a36 modulates pluripotency of mouse embryonic stem cells by regulating mitochondrial function and glutathione level. Biochem. J 476, 1585–1604. [DOI] [PubMed] [Google Scholar]
- 77.Lee SH, and McCormick F (2006). p97/DAP5 is a ribosome-associated factor that facilitates protein synthesis and cell proliferation by modulating the synthesis of cell cycle proteins. EMBO J. 25, 4008–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sugiyama H, Takahashi K, Yamamoto T, Iwasaki M, Narita M, Nakamura M, Rand TA, Nakagawa M, Watanabe A, and Yamanaka S (2017). Nat1 promotes translation of specific proteins that induce differentiation of mouse embryonic stem cells. Proc. Natl. Acad. Sci. USAUSA 114, 340–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smirnova VV, Shestakova ED, Nogina DS, Mishchenko PA, Prikazchikova TA, Zatsepin TS, Kulakovskiy IV, Shatsky IN, and Terenin IM (2022). Ribosomal leaky scanning through a translated uORF requires eIF4G2. Nucleic Acids Res. 50, 1111–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, and Sabatini DM (2012). A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. j 17, 10–12. [Google Scholar]
- 82.Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smith T, Heger A, and Sudbery I (2017). UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 27, 491–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kiniry SJ, Judge CE, Michel AM, and Baranov PV (2021). Trips-Viz: an environment for the analysis of public and user-generated ribosome profiling data. Nucleic Acids Res. 49, W662–W670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, and Hofacker IL (2011). ViennaRNA Package 2.0. Algorithm Mol. Biol 6, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Eng JK, Jahan TA, and Hoopmann MR (2013). Comet: an open-source MS/MS sequence database search tool. Proteomics 13, 22–24. [DOI] [PubMed] [Google Scholar]
- 87.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villén J, Haas W, Sowa ME, and Gygi SP (2010). A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Elias JE, and Gygi SP (2007). Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214. [DOI] [PubMed] [Google Scholar]
- 89.Elias JE, and Gygi SP (2010). Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol. Biol 604, 55–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Beausoleil SA, Villén J, Gerber SA, Rush J, and Gygi SP (2006). A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol 24, 1285–1292. [DOI] [PubMed] [Google Scholar]
- 91.Czechanski A, Byers C, Greenstein I, Schrode N, Donahue LR, Hadjantonakis AK, and Reinholdt LG (2014). Derivation and characterization of mouse embryonic stem cells from permissive and nonpermissive strains. Nat. Protoc 9, 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Darnell AM, Subramaniam AR, and O’Shea EK (2018). Translational Control through Differential Ribosome Pausing during Amino Acid Limitation in Mammalian Cells. Mol. Cell 71, 229–243.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, and Weissman JS (2012). The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc 7, 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, et al. (2019). GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li H, and Durbin R (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, and Carey VJ (2013). Software for computing and annotating genomic ranges. PLoS Comput. Biol 9, e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ting L, Rad R, Gygi SP, and Haas W (2011). MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods 8, 937–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McAlister GC, Nusinow DP, Jedrychowski MP, Wühr M, Huttlin EL, Erickson BK, Rad R, Haas W, and Gygi SP (2014). MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem 86, 7150–7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Paulo JA, O’Connell JD, and Gygi SP (2016). A Triple Knockout (TKO) Proteomics Standard for Diagnosing Ion Interference in Isobaric Labeling Experiments. J. Am. Soc. Mass Spectrom 27, 1620–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McAlister GC, Huttlin EL, Haas W, Ting L, Jedrychowski MP, Rogers JC, Kuhn K, Pike I, Grothe RA, Blethrow JD, and Gygi SP (2012). Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal. Chem 84, 7469–7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Ribo-seq data are available in NCBI’s Gene Expression Omnibus (GEO) through GEO Series accession number GEO: GSE248729 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE248729).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.