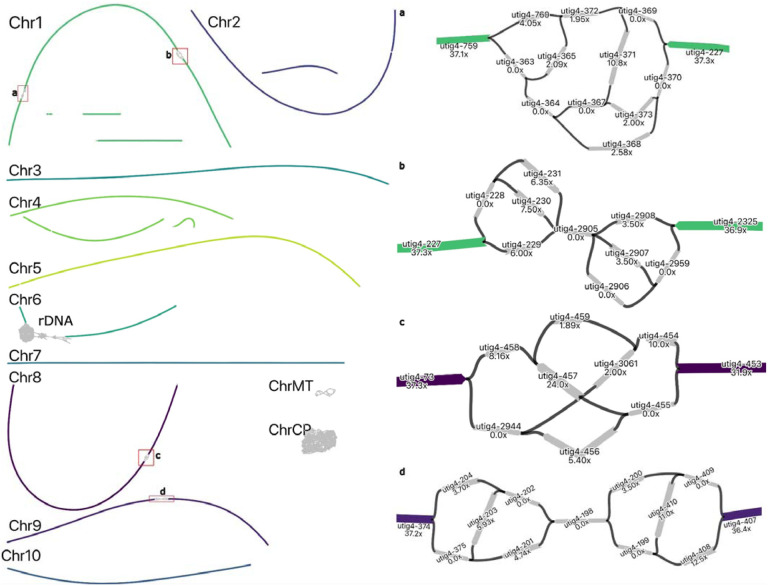
Figure 5: Duplex + ultra-long assembly graph for Z. mays prior to manual resolution.
In the maize assembly graph, most chromosomes are linear and resolved, except for regions of unresolved repeats (highlighted in red boxes), gaps on Chromosomes 1, 2, and 4, and the 45S rDNA array on Chromosome 6. ChrCP denotes the chloroplast and ChrMT denotes the mitochondria genomes, respectively. The callouts show detailed unresolved graph structure, as in Figure 4. All tangles are due to regions with low-coverage Duplex sequencing, which were gap-filled with ONT UL sequences. In each case, the path agreeing with the majority of ONT UL read alignments was selected for resolving the tangle. One end of Chromosome 3 was missing a telomere which was incorporated using ONT UL read consensus.