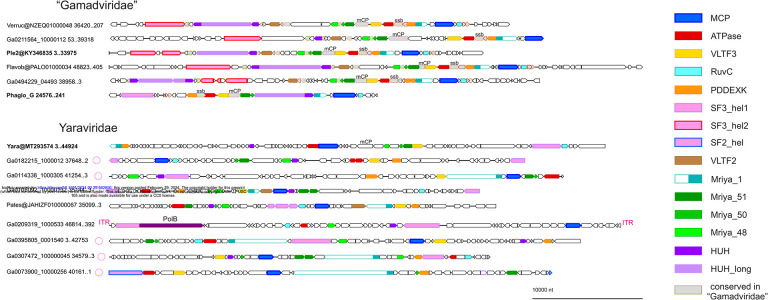
Figure 2. Genome maps of selected mriyaviruses.
Genes with predicted functions are shown by color-coded block arrows. Circles near contig names indicate contigs with direct terminal repeats. Abbreviations: ITR, inverted terminal repeats; PolB, family B DNA polymerase; mCP, minor capsid protein; ssb, single strand DNA binding protein; MCP, Major Capsid Protein; ATPase, DNA packaging ATPase; VLTF3, virus late transcription factor 3, RuvC, RuvC-like Holliday junction resolvase homologous to poxvirus A22 resolvase; PDDEXK, PDDEXK superfamily endonuclease; VLTF2, virus late transcription factor 2; Mriya_1, conserved domain homologous to Yaravirus gene 1; Mriya_51, Yaravirus gene 51 homolog; Mriya_50, Yaravirus gene 50 homolog; Mriya_48, Yaravirus gene 48 homolog; HUH, mriyavirus HUH endonuclease; HUH_long, conserved gamadvirus protein containing a C-terminal domain homologous to mriyavirus HUH endonuclease. Genome maps of all 60 mriyavirus representative genomes are available at https://ftp.ncbi.nih.gov/pub/yutinn/mriya_2024.