Figure 5: Alternative ERACR versus MCR-GFP outcomes.
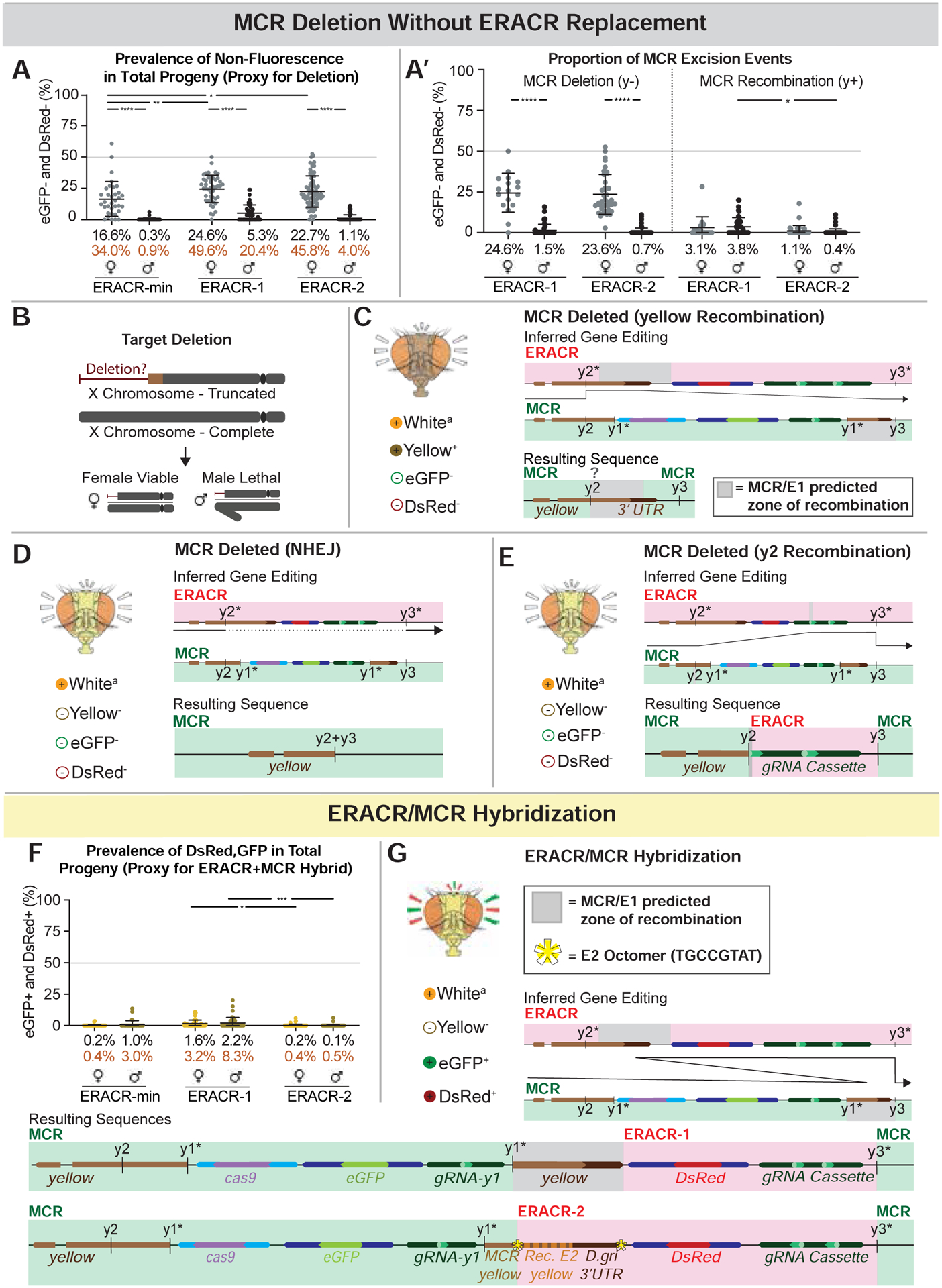
Partial copying or fusion of sequences on the receiver chromosome. A) Non-fluorescent DsRed−,GFP− F2 progeny as a proxy for MCR-GFP deletion events. A’) Data in panel A re-plotted by inheritance of y− (lethal deletion) versus y+ (recombination) alleles. Body color permits inference of the type of excision event that occurred, as depicted in C-E. B) Loss of essential sequences distal to the ERACR gRNA cut sites result in male-lethal alleles. Viability of several such alleles can be rescued in males by duplications covering the tip of the X chromosome (Fig. S12B). C-E) The MCR-GFP element is deleted, but not replaced by ERACR, producing three distinct observed outcomes. C) Hypothesized repair mediated by partial pairing of un-recoded y sequences carried by ERACR-1 and endogenous y sequences 3’ to the MCR-GFP element result in expression of the recoded y cassette and a wild-type body color. D) NHEJ events joining adjacent sequences at the gRNA-y2 and gRNA-y3 cut sites. E) Likely pairing between 17 bp of the gRNA-y2 genomic target sequences 5’ to the MCR-GFP with correspondingly oriented sequences in the gRNA-y2 transgene carried by either ERACR-min or ERACR-1. F) Prevalence of DsRed+, GFP+ F2 progeny in which MCR-GFP and ERACR sequences are both present on the receiver chromosome. G) Two examples of MCR-GFP/ERACR fusion events (see Fig. S14).
