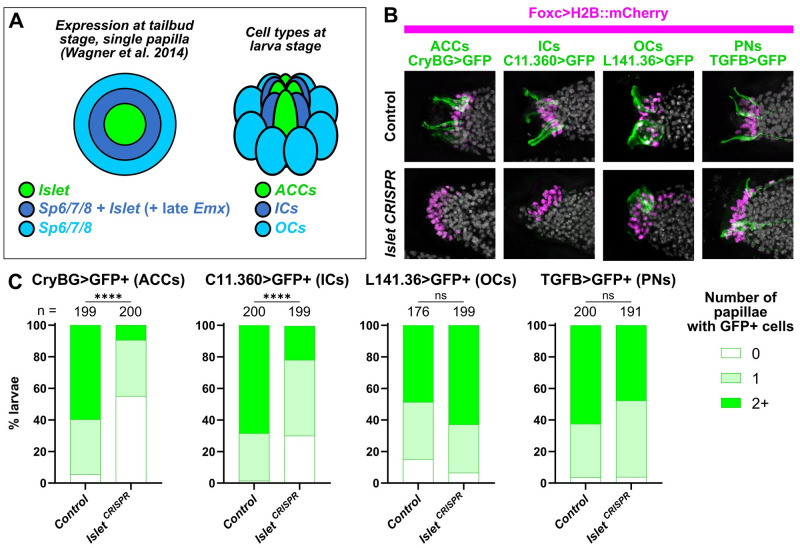
Fig 3. The transcription factor Islet is required for specification of ACCs and ICs.
(A) Diagram depicting a partially overlapping expression patterns of Islet and Sp6/7/8, as originally shown by in situ mRNA hybridizations (Wagner and colleagues), and the correlation of these patterns with the later arrangement of ACCs, ICs, and OCs in the papillae. “Late” Emx expression in a ring of cells expressing both Islet and Sp6/7/8 appears to be distinct from earlier Emx expression in Foxg-negative cells (see text and S2 Fig for details). (B) Papilla lineage-specific CRISPR/Cas9-mediated mutagenesis of Islet using Foxc>Cas9 and a the U6>Islet.2 sgRNA plasmid shows reduction of larvae showing expression of reporters labeling ACCs and ICs, but not OCs or PNs. Results compared to a negative “control” condition using a negative control sgRNA (U6>Control, see text for details). Nuclei counterstained with DAPI (white). (C) Scoring data for larvae represented in panel B, averaged between biological duplicates. Foxc>H2B::mCherry+ larvae were scored for quantity of papillae showing visible expression of the corresponding GFP reporter plasmid. Due to mosaic uptake or retention of the plasmids after electroporation, number of papillae with GFP fluorescence is variable and rarely seen in all 3 papillae even in control larvae. Normally larvae have 3 papilla (GFP+ or not), but some mutants have more/fewer than 3. ACC/IC/OC subpanels in panel B at 20 hpf/20 °C (~st. 29), PN subpanels at 21 hpf/20 °C (~st. 29). Same applies to scoring data in Panel C. All experiments were performed in duplicate, with number of embryos ranging from 76 to 100 per condition per replicate. **** p < 0.0001 in both duplicates, as determined by chi-square test. ns = not statistically significant in at least 1 duplicate, also by chi-square test. See S4 Data for the data underlying the graphs and for statistical test details. ACC, axial columnar cell; IC, inner collocyte; OC, outer collocyte; PN, papilla neuron; sgRNA, single-chain guide RNA.