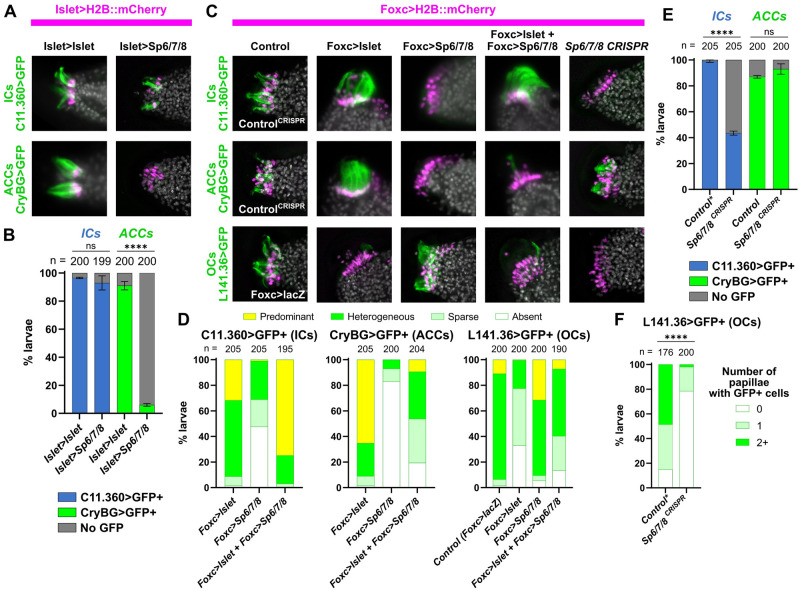
Fig 4. Specification of ACCs, ICs, and OCs by a combinatorial logic of Islet and Sp6/7/8.
(A) Overexpression of Sp6/7/8 (using the Islet>Sp6/7/8 plasmid) in all Islet+ papilla cells results in loss of ACCs (assayed by expression of CryBG>Unc-76::GFP, green), but not of ICs (assayed by expression of C11.360>Unc-76::GFP, green). Islet overexpression (with Islet>Flag::Islet-rescue) does not significantly impact the specification of ACCs or ICs. Larvae at 20 hpf/20 °C (~st. 29). (B) Scoring data showing presence or absence of ICs or ACCs in Foxc>H2B::mCherry+ larvae, as represented in panel A. Experiments were performed in duplicate with 99 or 100 larvae in each duplicate. (C) Cell type specification assayed by reporter plasmid expression (green) in larvae subjected to various Islet and/or Sp6/7/8 perturbation conditions (see main text for details). For ICs and ACCs, the “control” condition is negative control CRISPR (U6>Control), while for OCs it is Foxc>lacZ. Overexpression ACC/IC subpanels are at 18.5 hpf/20 °C (~st. 28), all CRISPR and OC panels at 20 hpf/20 °C (~st. 29). (D) Scoring data for most larvae represented in panel C. Foxc>H2B::mCherry+ larvae were scored for cell type-specific GFP reporter expression that was “heterogeneous” (mixed on/off GFP expression, with all “wild type” patterns of expression falling under this category), “predominant” (ectopic/supernumerary GFP+ cells), “sparse” (reduced frequency/intensity of GFP expression), or “absent” (no GFP visible). (E) IC or ACC reporter (C11.360>Unc-76::GFP, CryBG>Unc-76::GFP) expression scored in Foxc>H2B::mCherry+ larvae represented in top 2 panels of right-most column in C. Experiment was performed and scored in duplicate, with number of larvae in each duplicate ranging from 100 to 105. (F) OC-specific reporter (L141.36>Unc-76::GFP) expression scored in Foxc>H2B::mCherry+ larvae represented by the bottom/right-most subpanel in panel B. Scoring strategy same as in Fig 3. Asterisk denotes when a duplicate of the negative control condition was also used for plots in Fig 3, as multiple CRISPR experiments were performed in parallel. All experiments were performed in duplicate, with number of embryos ranging from 76 to 100 per duplicate. Foxc>Cas9 used for all CRISPR/Cas9 experiments. The Islet cis-regulatory sequence used (panels A and B) was always Islet intron 1 + -473/-9. For overexpression conditions, Foxc>lacZ or Islet>LacZ were used to normalize total amount of DNA (see S1 File for detailed electroporation recipes). All error bars indicate upper and lower limits. **** p < 0.0001 in both duplicates as determined by Fisher’s exact test (panels B and E) or chi-square test (panel F). ns = not statistically significant in at least 1 duplicate. See S4 Data for the data underlying the graphs and statistical test details. ACC, axial columnar cell; IC, inner collocyte; OC, outer collocyte.