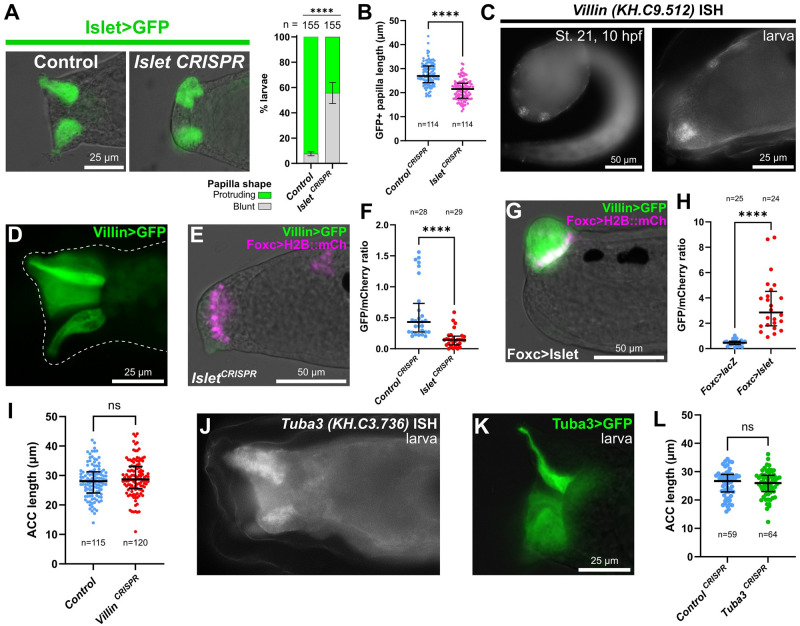
Fig 7. Islet is also required for papilla morphogenesis.
(A) Papilla shape is shortened and blunt at the apical end upon tissue-specific CRISPR/Cas9-mediated mutagenesis of Islet. Embryos were electroporated with Islet intron 1 + -473/-9>Unc-76::GFP and Foxc>Cas9. Islet CRISPR was performed using U6>Islet.2 sgRNA plasmid and the negative control used U6>Control. Larvae were imaged at 20 hpf/20 °C (~st. 28). Right: Scoring of percentage of GFP+ larvae classified as having normal “protruding” or blunt papillae, as represented to the left. Experiment was performed and scored in duplicate, using 2 different GFP fusions: Unc-76::GFP and DcxΔC::GFP [88]. Replicate 1: n = 100 for either condition; replicate 2: n = 55 for either condition **** p < 0.0001 in both duplicates as determined by Fisher’s exact test. (B) Quantification of papilla cell (Islet intron 1 +-473/-9>Unc-76::GFP+) lengths along apical-basal axis in negative control and Islet CRISPR larvae at 18 hpf/20 °C (~st. 28). Both Islet.1 and Islet.2 sgRNAs used in combination. Statistical significance tested by unpaired t test (two-tailed). See S8 Fig for duplicate experiment. (C) In situ mRNA hybridization of Villin, showing expression in Foxg+/Islet+ central papilla cells at 10 hpf/20 °C (st. 21, left) and at larval stage (~st. 27, right). (D) Villin -1978/-1>Unc-76::GFP showing expression in the papilla territory of electroporated larvae (~st. 28), strongest in the central cells. (E) Villin -1978/-1>Unc-76::GFP in st. 28 larvae is down-regulated by tissue-specific CRISPR/Cas9 mutagenesis of Islet (Foxc>Cas9 + U6>Islet.1 + U6>Islet.2, see text for details). (F) Quantification of effect of Islet CRISPR (as in panel E) on Villin -1978/-1>Unc 76::GFP/Foxc>H2B::mCherry mean fluorescence intensity ratios in ROIs defined by the mCherry+ nuclei (see Methods for details). Significance determined by Mann–Whitney test (two-tailed). (G) Villin reporter is up-regulated in st. 28 larvae by overexpressing Islet (Foxc>Islet, see text for details). (H) GFP/mCherry ratio quantification done in identical manner as in F, but comparing Islet overexpression (as in panel G) and control lacZ larvae. (I) Quantification of ACC lengths measured in negative control and papilla-specific Villin CRISPR larvae at 17 hpf/20 °C (~st. 27). Significance tested by unpaired t test (two-tailed). Although no statistically significant difference between control and CRISPR larvae was observed in this replicate, average ACC length was significantly shorter in the CRISPR condition in an additional replicate (S8 Fig). (J) mRNA in situ hybridization for Tuba3, showing enrichment in the central cells of the papillae in st. 27 larvae. (K) Tuba3>Unc-76::GFP reporter plasmid is broadly expressed in the papillae of st. 27 larvae but stronger in central cells. (L) Papilla-specific CRISPR knockout of Tuba3 does not result in decrease of average ACC apical-basal cell length compared to negative control CRISPR using U6>Control sgRNA instead. Significance tested by unpaired t test (two-tailed). ns = not significant. All large bars indicate medians and smaller bars indicate interquartile ranges. See S3 and S4 Data for the data underlying the graphs and for statistical test details. ACC, axial columnar cell; ROI, region of interest; sgRNA, single-chain guide RNA.