Summary
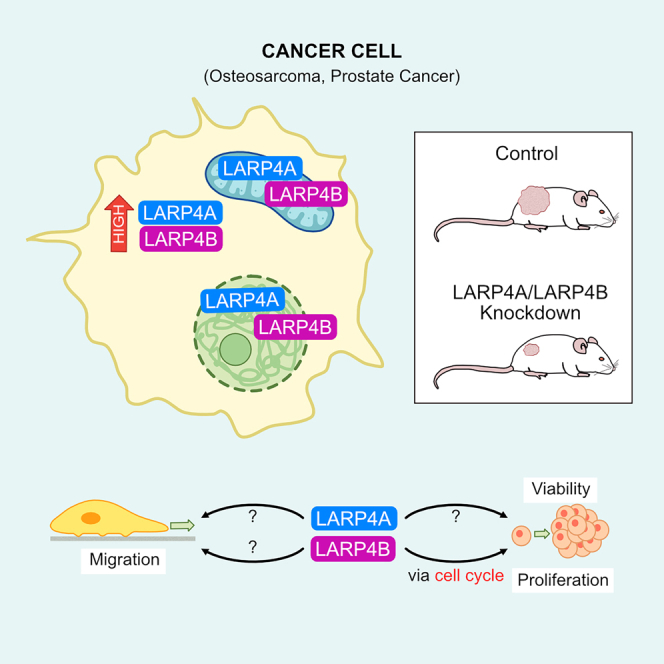
RNA-binding proteins (RBPs) are emerging as important regulators of cancer pathogenesis. We reveal that the RBPs LARP4A and LARP4B are differentially overexpressed in osteosarcoma and osteosarcoma lung metastases, as well as in prostate cancer. Depletion of LARP4A and LARP4B reduced tumor growth and metastatic spread in xenografts, as well as inhibiting cell proliferation, motility, and migration. Transcriptomic profiling and high-content multiparametric analyses unveiled a central role for LARP4B, but not LARP4A, in regulating cell cycle progression in osteosarcoma and prostate cancer cells, potentially through modulating key cell cycle proteins such as Cyclins B1 and E2, Aurora B, and E2F1. This first systematic comparison between LARP4A and LARP4B assigns new pro-tumorigenic functions to LARP4A and LARP4B in bone and prostate cancer, highlighting their similarities while also indicating distinct functional differences. Uncovering clear biological roles for these paralogous proteins provides new avenues for identifying tissue-specific targets and potential druggable intervention.
Subject areas: Natural sciences, Biological sciences, Biochemistry, Cell biology, Systems biology, Cancer systems biology
Graphical abstract
Highlights
-
•
LARP4A and LARP4B are overexpressed in prostate cancer and osteosarcoma
-
•
Knockdown of LARP4A and LARP4B reduces proliferation and tumorigenesis
-
•
Both LARP4 proteins regulate morphology and migration; LARP4B modulates cell cycle
-
•
LARP4A and LARP4B localize to mitochondria and nucleus in addition to cytosol
Natural sciences; Biological sciences; Biochemistry; Cell biology; Systems biology; Cancer systems biology
Introduction
Cancer is a heterogeneous disease in which a combination of genetic mutations and epigenetic changes enable cancer cells to divide more frequently than surrounding non-cancer cells, avoid cell death, and spread to secondary sites as metastases.1,2 Changes in gene expression are central to the development, progression, and spread of neoplastic disease. RNA binding proteins (RBPs) play central roles in this process, orchestrating the function and fate of coding and non-coding RNA molecules in a myriad of ways, including control of RNA splicing, polyadenylation, stability, localization, transport, translation, processing, metabolism, and decay.3,4,5,6 Growing evidence shows that RBPs are often dysregulated in cancer: their altered expression and/or mutations, as well as perturbations of RBP-RNA networks, have been causally associated with cancer onset, progression, and metastasis,7,8,9,10 but little is known of the mechanistic bases underlying these effects.
The La-Related Proteins (LARPs) are an ancient superfamily of RBPs with fundamental roles in RNA biology.11 They are present in all eukaryotes and consist of ∼250 members, evolutionarily divided into 5 families (LARP1, LARP3, LARP4, LARP6, and LARP7). The hallmark of the LARPs is a conserved winged helix domain called the La motif (LaM) that, with a few exceptions, is associated with a downstream RNA-recognition motif (RRM1), together comprising the La-module.11,12,13 The La-module functions as a bipartite RNA recognition unit,14,15,16,17,18 albeit some exceptions have started to emerge.19,20 Beyond this, each LARP family is characterized by family specific domains and motifs implicated in RNA and/or protein partner interactions, using an assortment of binding modes and contributing to the diversification of functional profiles.11,13 Although much remains to be explored, common as well as individual functional traits across the LARP families have started to be delineated.11,13 Several LARPs have been associated with cancer,10,11,21 including the two human LARP4 paralogs, LARP4A and LARP4B, also referred to as LARP4 and LARP5, respectively.
LARP4A and LARP4B have been described as predominantly cytoplasmic proteins that share overall domain organization and display 74% sequence identity in their La-module and ∼40% throughout.11 They interact with the Poly(A) binding protein (PABPC1) via a PAM2w motif located in the N-terminal region and a less-well defined PABPC-binding motif (PBM) downstream of the RRM1.11,20,21,22,23,24,25 Their common features also include interaction with receptor for activated kinase C (RACK1), enhancement of translation and mRNA stability, association with ribosomes, and localization to stress granules.11,23,24,25,26
Beyond this, other functions of either LARP4A or LARP4B have been steadily emerging. For instance, LARP4A has been shown to exhibit the protection of mRNA 3′ poly(A) tail lengths and the associated mRNA stabilization,27,28 contribute to mRNA metabolism during the activation of T cells immune response,29 play roles in the gene regulatory network governed by metazoan miRNAs30 and affect local mRNA translation at the mitochondria level,31 while LARP4B appears to reduce cell and organ sizes in Drosophila melanogaster via dMyc regulation.32 These findings may possibly underline functional differences between LARP4A and LARP4B, thereby endorsing a sub- or neo-functionalization hypothesis for these two paralogs that have been maintained through evolution.33,34 However, notably, data remain patchy and limited, as investigations have not been pursued in parallel on both proteins, precluding a comprehensive comparative profiling of LARP4A and LARP4B’s functional capacities. To date, investigations of RNA targets and RNA binding preferences have revealed that LARP4A associates with poly(A) RNA primarily via the PAM2w motif in a manner that is mutually exclusive to PABPC MLLE interaction,20,24 whereas LARP4B binds AU-rich sequences in 3′ untranslated regions (UTRs) in mature mRNAs25 using a different molecular mechanism (M.R.C., unpublished). How these different RNA recognition capabilities translate to the function of LARP4A and LARP4B remains to be elucidated.
The roles of LARP4A and LARP4B in cancer biology are equally not well understood, but sporadic evidence is accumulating in line with the increasing interest in RNA binding proteins in cancer progression.10,35 We and others first described LARP4A as a regulator of cancer cell morphology in a RNAi Drosophila screen and were subsequently shown to affect cell migration in breast, prostate, and ovarian cancer cell lines, albeit with varying effects.36,37,38 Since then, LARP4A has often been coined a tumor suppressor; however its roles in tumorigenesis and cancer growth have not yet been elucidated. Work on LARP4B has revealed varying roles depending on tumor or tissue type: in glioma, LARP4B has tumor suppressive properties because of its growth-restrictive roles,39,40 whereas it possesses oncogenic growth-promoting effects in an acute myeloid leukemia (AML) mouse model.41 Data from the TCGA TARGET GTEx study have associated altered LARP4A and LARP4B mRNA expression with different cancer subsets and, intriguingly, the upregulation of LARP4B mRNA was suggested to be an independent risk factor for the prognosis of patients with liver cancer.42 LARP4A and/or LARP4B have also appeared in several transposon mutagenesis screens to identify new genes driving tumorigenesis for colorectal cancer,43 hepatocellular carcinoma,44 malignant peripheral nerve sheath tumors45 and metastatic medulloblastoma.46 Furthermore, the enrichment of nonsense mediated decay (NMD)-elicit mutations in LARP4B was revealed in hypermutated stomach adenocarcinoma.47
The increasing association between the LARP4 proteins and cancer suggests complex and context-dependent functional roles for LARP4A and LARP4B in tumor biology.35 In this study, we have determined the contributions of LARP4A and LARP4B to cancer progression using osteosarcoma and prostate cancer cells, representing two distinct cancer origins, mesenchymal and epithelial, respectively. Using loss- and gain-of-function approaches, we provide a systematic comparison of their similarities and differences with respect to their cellular, transcriptional, and biological effects in vivo and in vitro. We reveal the key roles of LARP4A and LARP4B in cancer progression and identify candidate mechanisms underlying these functions.
Results
LARP4A and LARP4B are overexpressed in cancer tissues
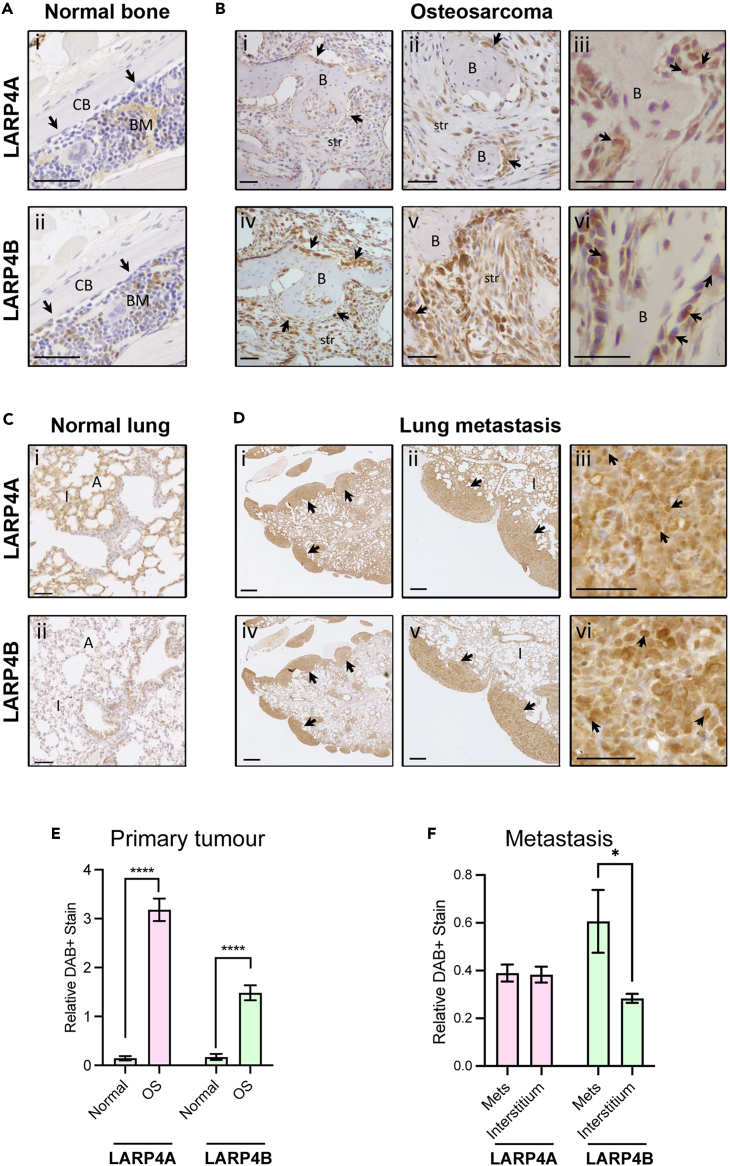
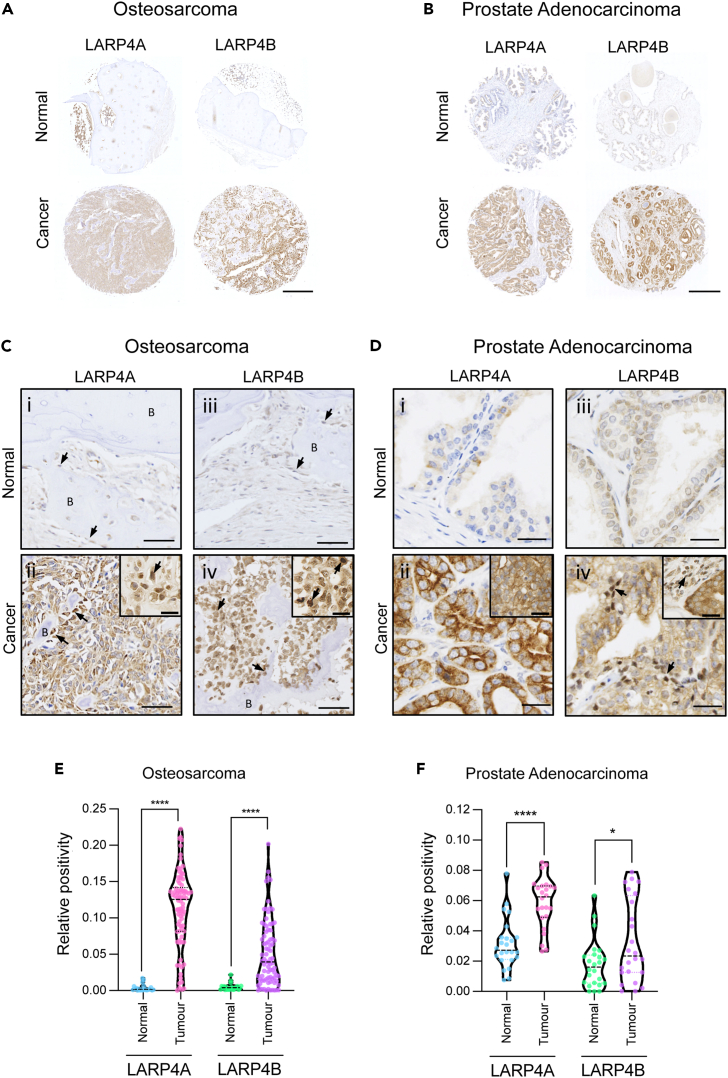
To understand the possible functions of LARP4A and LARP4B as oncoproteins or tumor suppressors in vivo, we used two approaches to investigate two cancer types that represent mesenchymal (sarcomas) and epithelial (carcinomas) tumors, namely osteosarcoma and prostate adenocarcinoma. In the first instance, we employed an established osteosarcoma transgenic mouse model48,49 to examine the expression of endogenous LARP4A and LARP4B expression in primary and metastastic osteosarcoma. Immunohistochemical staining of primary tumors revealed high expression of both LARP4A and LARP4B proteins in transformed osteoblasts lining neoplastic bone as well as within the intratumoral stroma, with negligible levels in normal bone (Figures 1A, 1B, and 1E). Further analysis of osteosarcoma lung metastasis49 detected high LARP4A expression within the normal pulmonary interstitium as well as in metastatic nodules (Figures 1Ci and 1DI,ii), whereas LARP4B displayed a striking profile with very low levels in normal lung interstitial cells and a relatively higher expression in metastatic foci (Figures 1C(ii), 1D(iv,v) and 1F). We further confirmed the expression of LARP4 proteins in human tumors using human tissue microarrays (TMAs). Immunohistochemical staining (Figures 2A–2D) and quantification (Figures 2E and 2F) demonstrated that both proteins are significantly overexpressed in both osteosarcoma and prostate adenocarcinoma compared to normal bone and prostate tissue, respectively (Figures 2A–2D), indicating that these RNA binding proteins are highly expressed in these cancers. Interestingly, nuclear staining for both LARP4A and LARP4B was evident in subpopulations of transformed osteoblasts and tumor stromal cells in murine primary osteosarcoma and lung metastases (Figures 1B(iii,vi) and 1D(iii,vi)), as well as in human transformed osteoblasts (Figures 2C(ii,iv)), and notably only LARP4B demonstrated a potential for nuclear localization in prostate cancer epithelial cells (Figures 2D(iv)). The high expression of LARP4 proteins in these cancers was endorsed following a pan-cancer analysis of LARP4A and LARP4B expression in patient tumor samples and normal tissues using the TCGA TARGET GTEx database,50 which showed that LARP4A and LARP4B mRNA are significantly overexpressed in the majority (60% and 52% respectively) of the 23 human cancer subsets, and significantly underexpressed in 20% and 32% of these compared to normal tissue controls (Figure S1A). Interestingly, both proteins were significantly upregulated in prostate adenocarcinoma tissue, although a lack of normal sample availability prevented the accurate quantification of mRNA expression in bone/sarcoma tissues using TCGA TARGET GTEx (Figures S1C and S1D).
Figure 1.
Expression of LARP4A and LARP4B in murine primary osteosarcoma and metastasis
(A) Immunohistochemical analysis of LARP4A and LARP4B protein expression in normal murine bone. Control bones show little to no expression of either LARP4A (A(i)) or LARP4B (A(ii)) in osteoblasts (arrows). CB - cortical bone (normal bone); BM – bone marrow. Scale bars, 50μm.
(B) Immunohistochemical analysis of LARP4A and LARP4B protein expression in a primary murine osteosarcoma.46,47 Increasing magnification of osteosarcoma tissues stained with LARP4A (B(i-iii)) or LARP4B (B(iv-vi)) show higher overall expression of LARP4B in transformed osteoblasts (arrows) and tumor stromal cells (str) compared to LARP4A. Arrows in B(iii) and B(vi) indicate nuclear staining of LARP4A and LARP4B, respectively. B – neoplastic bone (tumor). Scale bars, 50μm.
(C) Immunohistochemical analysis of LARP4A and LARP4B protein expression in normal murine lung tissue. Control lungs show markedly higher expression of LARP4A (C(i)) in pneumocytes of the lung interstitium compared to LARP4B (C(ii)). Scale bars, 200μm.
(D) Immunohistochemical analysis of LARP4A and LARP4B protein expression in lungs harboring osteosarcoma metastatic nodules following tail vein injection. Increasing magnification of lungs stained with LARP4A (D(i-iii)) or LARP4B (D(iv-vi)) show high expression in metastatic nodules (arrows). Arrows in D(iii) and D(vi) indicate nuclear staining of LARP4A and LARP4B, respectively. I – lung interstitium; A – alveolar sacs. Scale bars, (D(ii,v)) 200μm; (D(i,iii,iv,vi) 500μm. Immunohistochemistry staining in (A-D) is representative of tissues from 3 independent mice.
(E and F) Quantification of LARP4A and LARP4B expression in (E) primary murine osteosarcoma and (F) metastatic lung nodules. The data represent mean (±SEM) DAB-positive staining relative to hematoxylin staining in normal vs. osteosarcoma (OS) tissues (E) or lung interstitium vs. metastatic lung nodules (F) from three independent experiments. T-tests were used for testing statistical significance between normal vs. OS or interstitium vs. metastatic nodules per LARP4 stain; ∗p < 0.05; ∗∗∗∗p < 0.0001.
Figure 2.
Tissue microarray (TMA) analysis of LARP4A and LARP4B expression in human osteosarcoma and prostate adenocarcinoma
(A and B) Immunohistochemical analysis of LARP4A and LARP4B protein expression in representative core samples of (A) osteosarcoma and (B) prostate adenocarcinoma biopsies showed high expression of each LARP4 paralog in respective tumor tissues (Cancer) compared to Normal samples. Scale bars, 0.5mm.
(C) Histological analysis of representative human osteosarcoma sections showing little to no expression of either LARP4A (C(i)) or LARP4B (C(iii) in normal osteoblasts (arrows) lining bone (B), and markedly higher expression in tumor tissue (C(ii,iv)) particularly in transformed osteoblasts (arrows). Insets in C(ii) and C(iv) depict the nuclear localization of LARP4A and LARP4B, respectively (arrows). Scale bars, 50um; insets 25um.
(D) Histological analysis of representative prostate adenocarcinoma sections showing little to no expression of either LARP4A D(i) or LARP4B (iii) in normal prostate tissue, and markedly high expression in transformed prostate epithelial cells in tumor tissue (D(ii,iv). Arrows in D(iv) and inset depict the nuclear localization of LARP4B. Scale bars, 25um.
(E and F) Quantification of LARP4A and LARP4B expression in (E) osteosarcoma and (F) prostate adenocarcinoma TMAs. The data represent the mean (±SEM) area of DAB-positive staining following LARP4A/LARP4B staining in normal and cancer tissues relative to the overall core area. For osteosarcoma, n = 11 duplicate cores for normal bone, n = 81 duplicate cores for cancer tissue, areas of bone marrow was excluded from analyses. For prostate adenocarcinoma, n = 12 duplicate cores for each normal and cancer tissues. T-tests were used to test for statistical significance between normal vs. cancerous tissues for each cancer type and LARP4 stain. ∗p < 0.05; ∗∗∗∗p < 0.0001.
Depletion of LARP4A and LARP4B markedly inhibits tumor growth in vivo
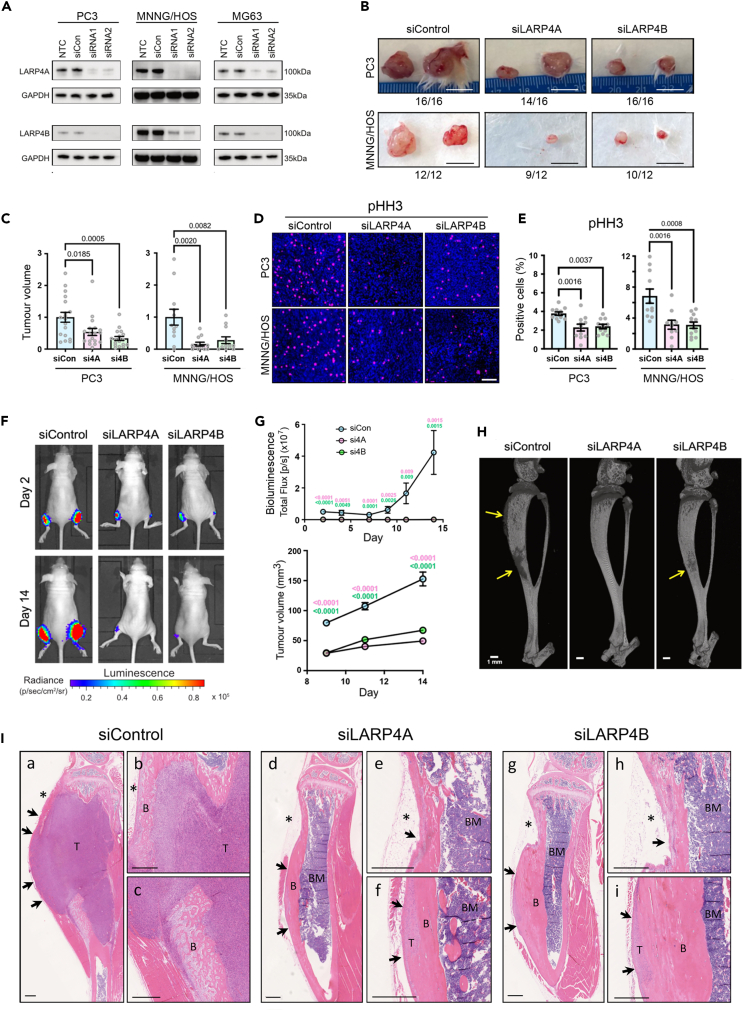
To investigate the functional consequences of LARP4A/LARP4B in cancer progression, we performed xenograft studies following genetic silencing of each protein in representative osteosarcoma (MNNG/HOS; MG63) and prostate cancer (PC3) cell lines. Western blot analysis indicated efficient silencing of LARP4A and LARP4B proteins using targeted siRNAs in PC3, MNNG/HOS, and MG63 cells (Figure 3A) which persisted for at least 7 days in vitro, and showed no evidence of paralog compensation (Figure S2). Subcutaneous injection of LARP4A/LARP4B-depleted PC3 and MNNG/HOS cells in immunocompromised mice showed a marked reduction in tumor volume by 50–85% in both PC3- and MNNG/HOS-derived xenografts (Figures 3B and 3C). Tumors formed by LARP4A and LARP4B-depleted cells had significantly lower expression of the mitosis and proliferation markers pHH3 and Ki67 while the expression of the apoptotic marker Cleaved Caspase-3 was not altered (Figures 3D, 3E, and S3A‒S3D), suggesting that LARP4A and LARP4B support prostate cancer and osteosarcoma tumor proliferation in vivo. We further assessed LARP4A and LARP4B tumorigenic capacities using a more biologically relevant orthotopic osteosarcoma xenograft model.49 Paratibial injection of MNNG/HOS cells depleted for either LARP4A or LARP4B resulted in a striking reduction in tumor formation compared to controls, as demonstrated by the low/undetectable luciferase luminescence signal over the experimental time-course and a significant decrease in tumor volume (Figures 3F and 3G). As MNNG/HOS cells are known to produce ectopic bone in tumor xenografts,51 we also monitored the bone-forming potential of LARP4A and LARP4B knockdown xenografts. While the presence of ectopic bone was confirmed by microCT analysis in siControl xenografts, this was completely absent or dramatically reduced in bones engrafted with siLARP4A- and siLARP4B-depleted cells, respectively (Figure 3H). Histological analysis demonstrated large tumor masses formed by siControl cells including ectopic bone, both of which were absent or hugely diminished in siLARP4A and siLARP4B xenografts (Figure 3I).
Figure 3.
Depletion of LARP4A and LARP4B inhibits tumor formation in vivo
(A) Western blot analysis of endogenous LARP4A and LARP4B expression in PC3, MNNG/HOS, and MG63 cells 72h following transfection with two siRNAs each targeting LARP4A (siRNA1 and siRNA2, top panel) or LARP4B (siRNA1 and siRNA2, bottom panel) as indicated. Non-transfected (NTC) and non-targeting siRNA (siCon) controls are shown and GAPDH was used as a loading control.
(B) Representative images of xenograft tumors formed following subcutaneous injection of PC3 (top) or MNNG/HOS (bottom) cells following transfection with non-targeting siRNA (siControl) or siRNA-mediated knockdown of LARP4A (siLARP4A) or LARP4B (siLARP4B). Numbers below each panel denote the proportion of the engraftment of each cell line after 2 weeks. Scale bars, 1cm.
(C) Quantification of volumes of subcutaneous tumors shown in (B) formed by siLARP4A and siLARP4B knockdown PC3 (left) and MNNG/HOS (right) cells, 2 weeks post-engraftment and normalized to siControl (siCon) tumors. The data represent the mean ± SEM, n = 6–8 mice per condition. p values are indicated.
(D) Representative immunofluorescence images of pHH3 and DAPI staining in subcutaneous tumors formed two weeks following the engraftment of siControl, siLARP4A, or siLARP4B PC3 or MNNG/HOS cells. Scale bar, 100μm.
(E) Quantification of the proportion of cells within PC3 (left) and MNNG/HOS (right) xenografts expressing pHH3 shown in (D), as a percentage of the total number of cells (DAPI). Bars represent the mean ± SEM per tumor, n = 9–16 individual tumors from 6 to 8 mice. p values are indicated.
(F) Representative bioluminescence images of paratibial xenografts of siControl, siLARP4A, or siLARP4B MNNG/HOS cells the day after and 14 days post-engraftment.
(G) Quantification of the bioluminescence signal (top) and palpable tumor volume (bottom) in mouse tibiae over a 14 days time course. Data represent mean ± SEM, two tibiae measured from n = 8 mice per control or knockdown condition. p values are indicated (pink value indicates p value of the siControl vs. siLARP4A comparison; green of the siControl vs. siLARP4B comparison).
(H) Representative microCT scans of murine tibiae following engraftment with control (siControl), LARP4A (siLARP4A), or LARP4B (siLARP4B) knockdown MNNG/HOS cells. Arrows highlight areas of malignant bone deposits as a result of osteosarcoma tumor formation. Scale bars, 1mm.
(I) Representative histological images of tibiae 14 days post-engraftment with siControl, siLARP4A, or siLARP4B knockdown MNNG/HOS cells. Low magnification images show a clear reduction in tumor mass in both siLARP4A (I(d)) and siLARP4B (I(g)) knockdown xenografts compared to siControls (I(a)). High magnification images in panels I(b) and I(c) (siControl), I(e) and I(f) (siLARP4A) and I(h) and I(i) (siLARP4B) show areas of tumor tissue (T) that are markedly reduced in the LARP4A and 4B knockout xenografts. B, Bone, T, Tumor, BM, Bone Marrow. Asterisks indicate the cell injection sites. Arrows indicate the tumor perimeter which is reduced in the knockout xenografts. Scale bars, 500μm. See also Figure S3.
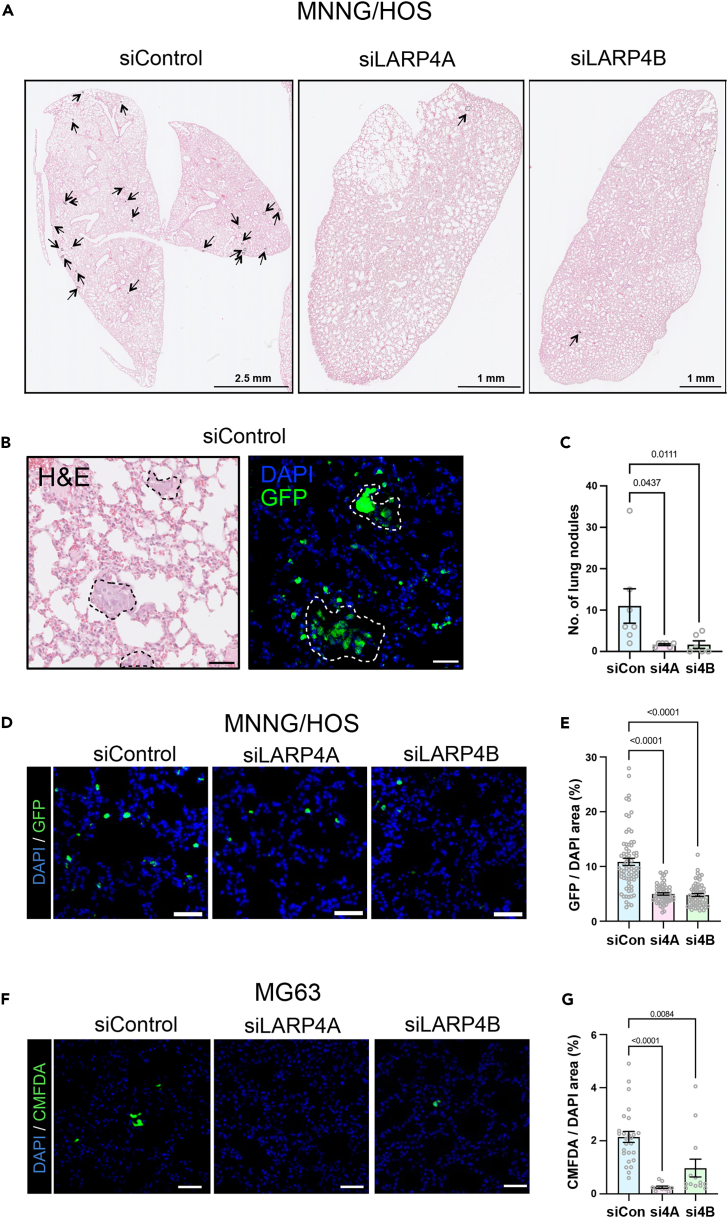
Finally, we assessed the metastatic potential of LARP4A and LARP4B-depleted osteosarcoma cells in vivo. Whereas siControl MNNG/HOS cells formed detectable nodules in the lungs within 7 days, LARP4A and LARP4B-depleted cells formed significantly less foci (Figures 4A–4C, and S4), with a concurrent, significant decrease in their ability to seed or expand in pulmonary tissue (Figures 4D and 4E). This was confirmed with the additional osteosarcoma cell line MG63, which demonstrated a significant decrease in cell engraftment as early as 24h post-injection (Figures 4F and 4G), indicating an essential role for these proteins in promoting metastatic colonization by tumor cells. Taken together, these in vivo expression and xenograft studies suggest clear pro-tumorigenic and metastatic roles for both LARP4A and LARP4B.
Figure 4.
Depletion of LARP4A and LARP4B inhibits lung metastasis
(A) Low magnification histological images (H&E) of lungs 7days following the tail vein injection of MNNG/HOS cells following transfection with non-targeting siRNA (siControl) or siRNA-mediated knockdown of LARP4A (siLARP4A) or LARP4B (siLARP4B). Black arrows indicate metastatic nodules. Scale bars are denoted within the images.
(B) High magnification of representative histological (H&E) and immunofluorescence (GFP/DAPI) images of MNNG/HOS GFP-positive metastatic nodules 7days following tail vein injection. Scale bar, 50μm.
(C) Quantification of the total number of metastatic nodules lesions per mouse 7days following the tail vein injection of control (siCon), LARP4A (si4A) or LARP4B (si4B) knockdown MNNG/HOS cells. The data represent the mean ± SEM, n = 6 mice per condition. p values are indicated.
(D) Representative immunofluorescence images of MNNG/HOS-GFP+ metastatic cells present in lung interstitium 7days following the tail vein injection of siControl, siLARP4A, or siLARP4B knockdown cells. Scale bars, 50μm.
(E) Quantification of the proportion of MNNG/HOS-GFP+ cells 7days following the tail vein injection of knockdown cells shown in (D). The data represent the mean ± SEM of GFP+ cells relative to DAPI+ cells from 10 randomly selected fields in n = 6 lungs per condition. p values are indicated.
(F) Representative immunofluorescence images of murine lungs 24h following the tail vein injection of CMFDA-labelled siControl, siLARP4A, or siLARP4B knockdown MG63 cells. Scale bars, 50μm.
(G) Quantification of the proportion of CMFDA-labelled MG63 cells 24h following the tail vein injection of knockdown cells shown in (F). The data represent the mean ± SEM, n = 3 mice per condition. p values are indicated. See also Figure S4.
LARP4A and LARP4B promote cell proliferation and LARP4B influences cell cycle progression
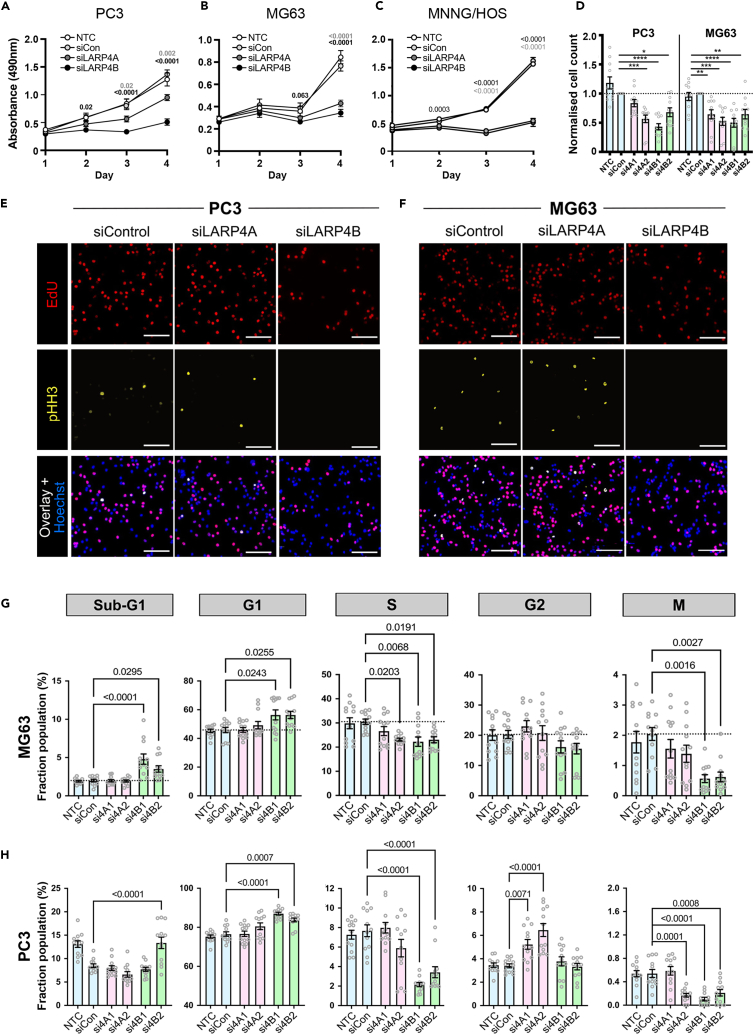
To uncover potential mechanisms underpinning the pro-tumorigenic roles of LARP4A and LARP4B in vivo, we compared the function of these paralogs on cell proliferation and viability. MTS assays demonstrated that the depletion of either LARP4A or LARP4B proteins by targeted siRNA silencing significantly reduced the viability of PC3, MG63, and MNNG/HOS cell lines compared to controls, and this was verified by the quantification of cell numbers (Figures 5A–5D), suggesting that both proteins positively regulate cell proliferation.
Figure 5.
LARP4A and LARP4B regulate cell proliferation and LARP4B affects cell cycle progression
(A‒C) MTS viability assays of PC3 (A), MG63 (B) and MNNG/HOS (C) control cells (non-transfected NTC, siCon) and siLARP4A or siLARP4B knockdown cells over a 4days time course. The data represent the mean ± SEM, n = 8 from 4 independent experiments. p values are indicated (light gray value indicates the p value of the siControl vs. siLARP4A comparison; black of the siControl vs. siLARP4B comparison).
(D) Quantification of PC3 (left) and MG63 (right) cell numbers after 72h following knockdown of siLARP4A (si4A1, si4A2), and siLARP4B (si4B1, si4B2) normalized to non-targeting siRNA (siCon) controls. NTC – non-transfected controls. The data represent the mean ± SEM, n = 11 from three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(E and F) Immunofluorescence images of siControl, siLARP4A, and siLARP4B knockdown PC3 (E) and MG63 (F) cells following EdU incorporation and pHH3 staining. Hoechst was used as a counterstain. Scale bars, 200μm.
(G and H) Multiparametric cell cycle analysis of PC3 (G) and MG63 (H) cells either non-transfected (NTC) or transfected with non-targeting control (siCon) and siRNAs targeting LARP4A (si4A1, si4A2), LARP4B (si4B1, si4B2) as indicated. The fraction of the population at each cell cycle stage (sub-G1, G1, S, G2, and M phases) for each cell type is depicted. The data represent the mean ± SEM, n = 12 from four independent experiments. p values are indicated. See also Figure S5.
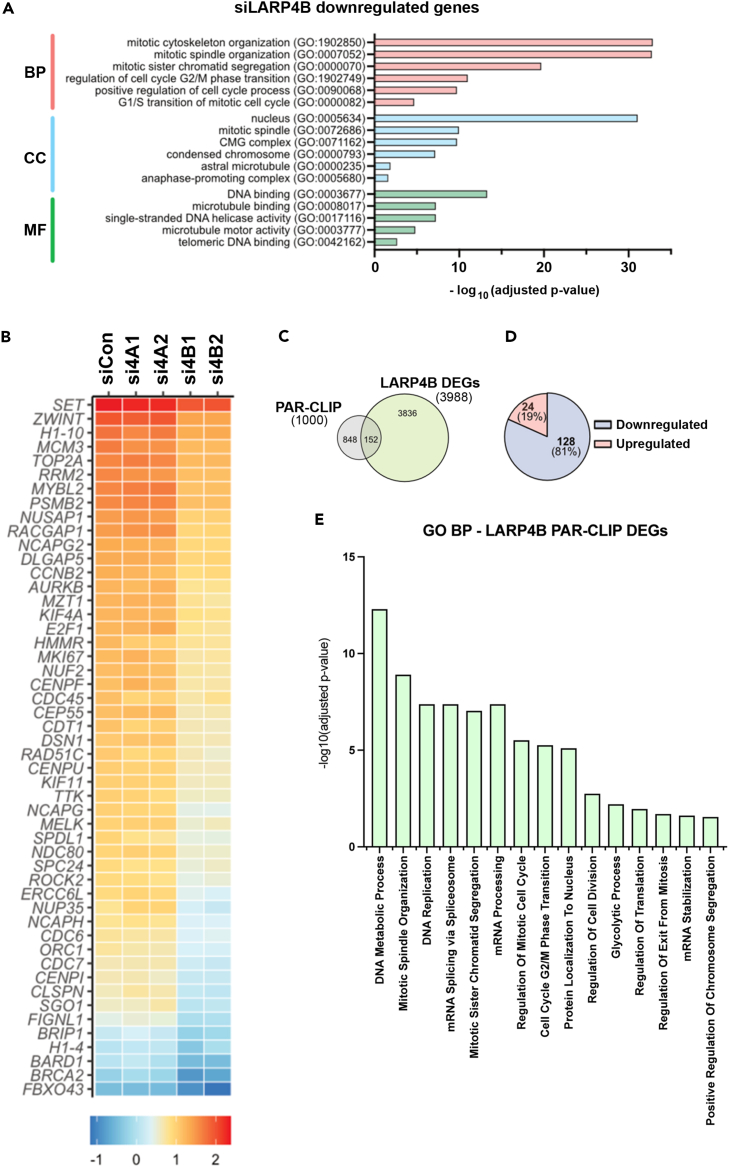
Based on reports that LARP4 proteins, in particular LARP4B, might perturb cell cycle,40,41 we investigated cell cycle distribution in LARP4A- and LARP4B-depleted MG63 and PC3 cells. High content multiparametric analysis52 of separate cell cycle stages showed that LARP4B knockdown in both cell lines significantly increased the fraction of cells in sub-G1 and G1 phases with a concomitant reduction in S and M phases (Figures 5E–5H), indicating that the depletion of LARP4B resulted in reduced cell cycle entry, slower progression through G1/G2 phases and decreased mitosis. In contrast, LARP4A depletion did not appreciably alter the cell cycle, with the minor inhibition of the S and M phases in MG63 cells (Figures 5E–5H). That LARP4B exhibited a greater effect than LARP4A on cell cycle was confirmed by overexpressing GFP-tagged proteins (Figures S5A and S5B). LARP4B overexpression increased the fraction of cells in S and M phases in both MG63 and PC3, whereas LARP4A overexpression had no effect (Figures S5C and S5D). LARP4B therefore enhances cell cycle progression, possibly by promoting cellular replication and mitosis. In agreement with this, RNAseq analysis revealed that the siRNA-depletion of LARP4B in MG63 and PC3 cells resulted in decreased RNA levels for genes over-represented in biological processes involved in the regulation of mitosis, spindle organization and cell cycle phase transition (Figures 6A and 6B). Specific genes include cyclins (CCNB1 and CCNE2), the positive cell-cycle regulator CDC7,53 the transcription factor E2F1 which promotes the expression of genes involved in S-phase entry and mitosis,54 the proliferation marker KI67, whose expression is controlled during cell cycle55 and BRCA2, the tightly regulated expression of which is highly critical in proliferation and cell cycle checkpoints (Figure 6B).56 Intriguingly, and in agreement with cell cycle multiparametric analysis, these changes were not observed in LARP4A-depleted cells (Figure 6B).
Figure 6.
Identification of LARP4A and LARP4B targets
(A) Gene ontology (GO) terms for Biological Process (BP), Cell Component (CC), and Molecular Function (MF) significantly enriched for, following bulk RNAseq analysis of significantly downregulated genes in LARP4B knockdown PC3 cells compared to siControl cells.
(B) Heatmap depicting relative expression (bottom scale Z score) of the top 50 mitosis-related genes across siControl (siCon), siLARP4A (si4A1, si4A2), and siLARP4B (si4B1, si4B2) knockdown PC3 cells.
(C) Venn diagram showing overlap between mRNAs that feature as differentially expressed genes (DEGs) between LARP4B knockdown and control cells in bulk RNAseq, and candidate direct mRNA targets (top 1000) of LARP4B as shown by PAR-CLIP analysis by Kuspert et al.25
(D) Pie chart demonstrating the proportion of upregulated and downregulated DEG/PAR-CLIP genes that overlap with bulk RNAseq analysis of LARP4B knockdown PC3 cells compared to controls.
(E) Gene ontology (GO) terms for 15 of the top biological processes (BP) significantly enriched for, following analysis of significantly downregulated putative mRNA target genes of LARP4B, in LARP4B knockdown PC3 cells compared to controls. See also Figure S6.
Cross-examination of mRNA abundance changes from our RNAseq with putative mRNA binding targets of LARP4B, identified in a previous PAR-CLIP study performed in HEK293 cells,25 revealed a subset of 152 cellular mRNAs that may be regulated directly by LARP4B at the steady-state level (Figure 6C). Of these, more than 80% were downregulated upon LARP4B depletion (128 genes; Figure 6D), implying that LARP4B might play a major role in stabilizing these mRNA targets,25 although this needs to be confirmed. Notably, these genes overwhelmingly enriched for many mitosis and cell cycle-related gene ontology (GO) terms (Figure 6E) and specifically include cyclins (CCNB1, CCNE2), cyclin-dependent kinases (CDK1/2) and their inhibitors (CDKN1A/B/C) and mitotic checkpoint kinases (AURKA/B). We confirmed the downregulation of several of these mitosis-related candidate targets of LARP4B to varying degrees in both PC3 and MG63 cells, namely AURKB, CCNB1, CCNE2, and E2F1 both at the mRNA and protein levels upon siRNA LARP4B depletion (Figures S6A‒S6D). Silencing of LARP4A also reduced the protein expression of E2F1, cyclin B1, cyclin E2, and AuroraB, but this was only in MG63 cells and the effects were markedly weaker than those following LARP4B knockdown. Thus, although LARP4A might regulate some phases of the cell cycle in a cell type-specific manner, LARP4B appears to be the prime regulator of genes involved in the cell cycle, in agreement with its role in the cell cycle and proliferation.
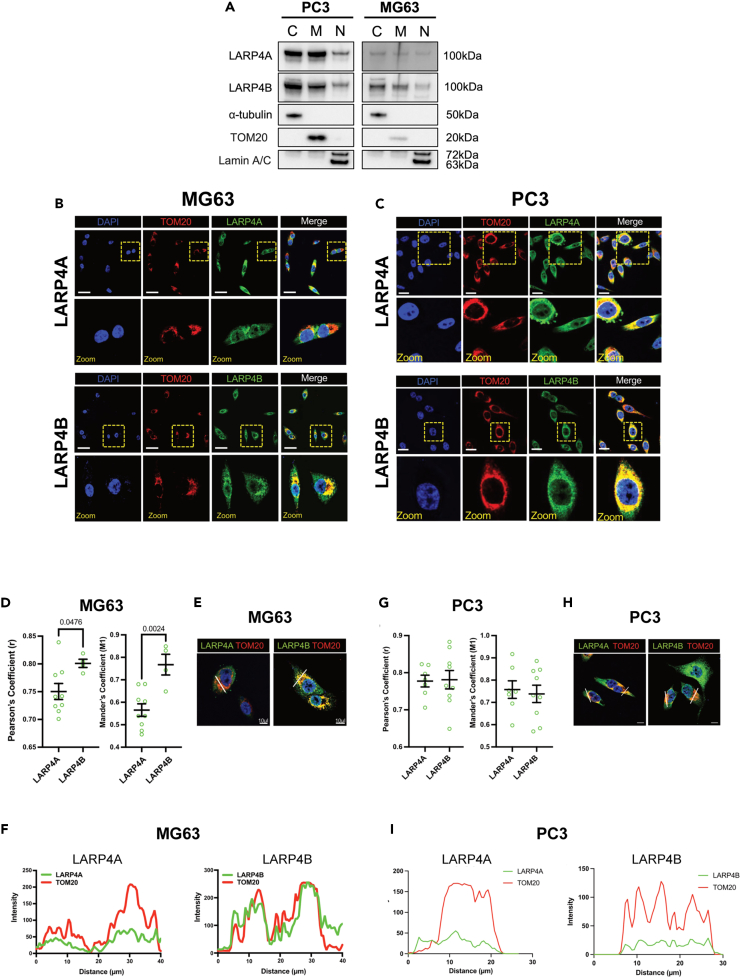
Recent work has reported that LARP4A localizes to mitochondria and regulates mitochondrial activity31,57 which may, at least in part, contribute to the cell proliferation phenotype observed. However, thus far no data are available for LARP4B. Our cell fractionation revealed that, besides the cytoplasm, LARP4A and LARP4B were expressed at comparable levels in mitochondrial fractions as well as in the nucleus to varying degrees in both MG63 and PC3 cells (Figure 7A). Immunofluorescence and confocal microscopy analyses corroborated these findings, demonstrating strong LARP4A and LARP4B expression within nuclear compartments both visually and following co-localization analyses with DAPI (Figure S7), as well as their expression alongside the mitochondrial marker, TOM20 (Figures 7B and 7C). Interestingly, in MG63 cells LARP4B showed a higher degree of co-localization with TOM20, compared to LARP4A (Figures 7D–7F), whereas there was a strong and largely equivalent co-localization of both LARP4A/TOM20 and LARP4B/TOM20 in PC3 cells (Figures 7G–7I). These data reveal mitochondrial localization for both proteins in both cell lines, which is in agreement with existing evidence of an important role for LARP4A in mitochondrial regulation31,57 and may suggest yet unidentified mitochondrial roles for LARP4B.
Figure 7.
Expression and localization of LARP4A and LARP4B in mitochondria
(A) Western blot analysis of endogenous LARP4A and LARP4B in PC3 and MG63 cells following the fractionation of cytoplasm (C), mitochondria (M) and nuclei (N). α-tubulin, TOM20, and Lamin A/C are used as respective controls to determine fraction purities.
(B and C) Immunofluorescence detection of TOM20 in (B) PC3 and (C) MG63 cells and co-localization with LARP4A and LARP4B proteins with DAPI counterstain. Scale bars, 25μm. Areas in yellow squares are depicted at higher magnification in lower panels.
(D) Pearson’s and Mander’s coefficient analyses showing a higher degree of LARP4B/TOM20 co-localization compared with LARP4A/TOM20 in MG63 cells. The data represent the mean ± SEM, n = 6 from three independent experiments. p values are indicated.
(E and F) Fluorescent intensity line plot profiling demonstrating the co-localization of LARP4A and LARP4B with the TOM20 mitochondrial marker in MG63 cells. White lines in (E) depict the areas used for LARP4A/TOM20 and LARP4B/TOM20 line plot analyses as indicated in (F). Scale bars in (E), 10μm.
(G) Pearson’s and Mander’s coefficient analyses showing a similar degree of LARP4B/TOM20 co-localization compared with LARP4A/TOM20 in PC3 cells. The data represent the mean ± SEM, n = 6 from three independent experiments. p values are indicated.
(H and I) Fluorescent intensity line plot profiling demonstrating the co-localization of LARP4A and LARP4B with the TOM20 mitochondrial marker in PC3 cells. White lines in (H) depict the areas used for LARP4A/TOM20 and LARP4B/TOM20 line plot analyses as indicated in (I). Scale bars in (H), 10μm. See also Figure S7.
Finally, we investigated whether apoptosis contributed to the reduction in prostate cancer and osteosarcoma cell number upon LARP4A/4B protein knockdown, given the previous suggestion that LARP4B modulates apoptosis in glioma cells.40 While a slight increase in the proportion of apoptotic cells was observed in LARP4A- and LARP4B-depleted cells compared to controls, this was not statistically significant (Figures S8A and S8B). Confirming these results, no change in apoptosis was detected in cells overexpressing LARP4A or LARP4B (Figures S8C and S8D). Notably, while mRNA levels of the pro-apoptotic factor BAX increased upon the overexpression of LARP4B in glioma cells,40 this was unaltered in LARP4A- or LARP4B-depleted PC3 or MG63 cells (Figures S8E and S8F). Moreover, RNAseq analysis showed that differentially expressed genes in LARP4A- or LARP4B-depleted cells compared to control did not enrich for processes involved in apoptosis, necrosis, and cell death with the parameters used, with no significant changes in mRNA abundance of key apoptotic regulators (e.g., BAD, BCL2, BID, MCL1, and BIK; Figure S9).
Taken together, loss- and gain-of-function studies suggest that LARP4A and LARP4B promote cancer cell proliferation and survival. In this context, this appears to be independent of apoptosis and, primarily for LARP4B, attributed to specific changes in cell cycle and mitosis regulation.
LARP4A and LARP4B induce morphological changes and promote cancer cell migration in vitro
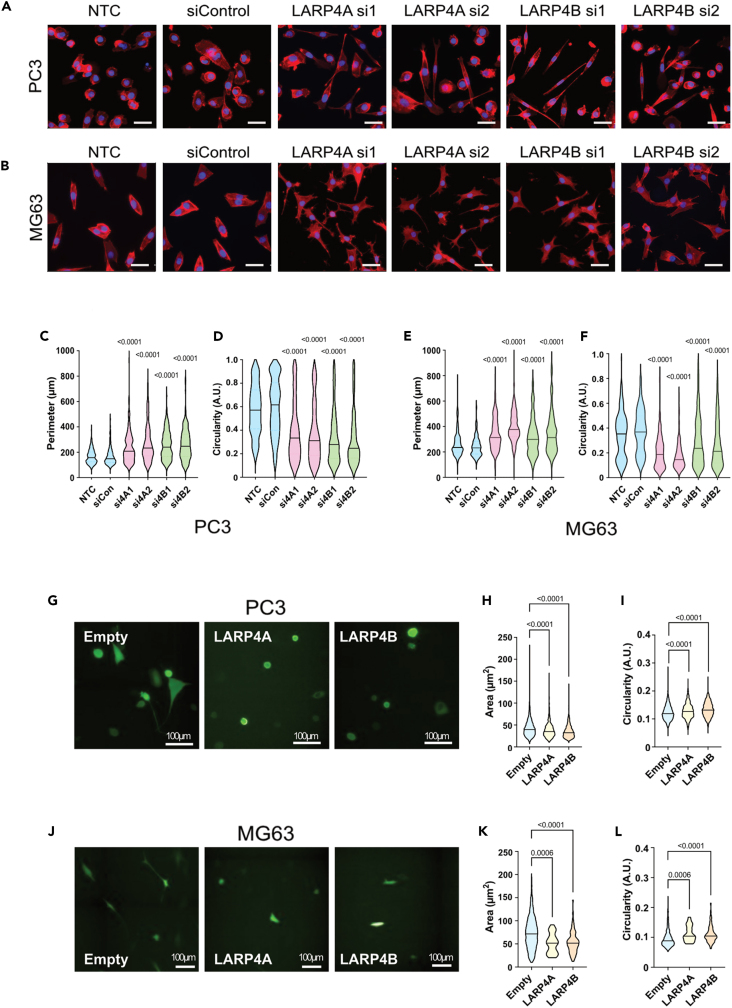
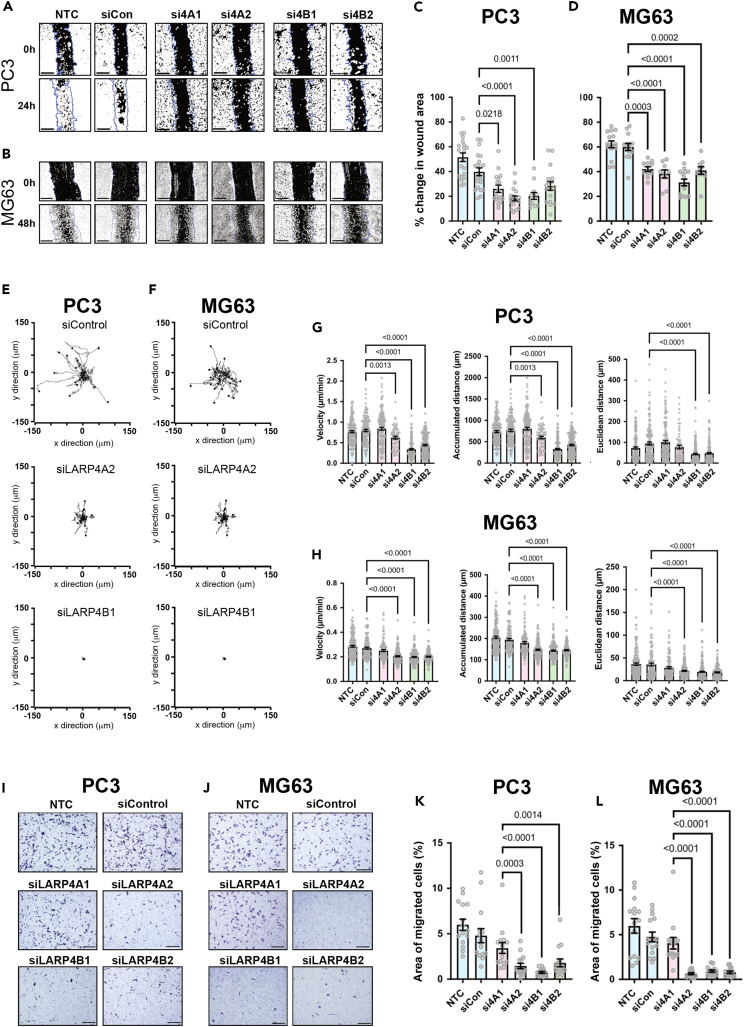
Morphological transformation and increased migration are well-established hallmarks of the transformed phenotype. LARP4A has previously been reported to regulate both cell morphology and migration,36,37,38 but LARP4B has not yet been explored in this context. Depletion of LARP4A or LARP4B in both PC3 and MG63 cells induced increased cell perimeter, elongation, and protrusion formation compared to controls (Figures 8A–8F). Conversely, the overexpression of LARP4A or LARP4B resulted in higher cell circularity and loss of protrusions in both cell types (Figures 8G‒8L), suggesting that these proteins promote cell rounding. To determine whether the observed cell morphological features may be due to differences in epithelial-to-mesenchymal transition (EMT), the expression of EMT biomarkers was examined. Neither depletion nor overexpression of either LARP4A or LARP4B significantly altered the expression of E-cadherin (PC3 cells) or vimentin (MG63 cells) (Figures S10A and S10B), indicating that these proteins are unlikely to act through EMT. We next determined whether these morphological alterations affected cell migration. LARP4A/4B-depleted PC3 and MG63 cells inhibited cell migration in a scratch assay (Figures 9A–9D). To rule out that a reduction in wound healing effects may, at least in part, arise from the reduced proliferation rates in LARP4A and LARP4B-depleted cells, we tracked the migratory paths of random, non-proliferating single cells over time. Consistent with previous results, the knockdown of LARP4A and LARP4B significantly reduced cell velocity, accumulated, and Euclidean distance in both PC3 and MG63 cell lines (Figures 9E–9H and Videos S1, S2, S3, S4, S5, S6, S7, S8, S9, S10, S11, and S12). Finally, transwell assays demonstrated that the silencing of LARP4A, and to a greater extent LARP4B, significantly impaired chemoattractant-mediated cell migration of all cell lines (Figures 9I‒9L and S11).
Figure 8.
LARP4A and LARP4B regulate cell morphology
(A and B) Immunofluorescence images of PC3 (A) or MG63 (B) control (NTC, siControl) and LARP4A (LARP4Asi1, LARP4Asi2) or LARP4B (LARP4Bsi1, LARP4Bsi2) knockdown cells stained with phalloidin and DAPI for morphological analyses. Scale bars, 50μm.
(C‒F) Morphological analysis of PC3 (C and D) and MG63 (E and F) cell perimeter and circularity as indicated. For all violin plots, the line depicts the mean, n = 178–220 cells from 3 independent experiments. p values are indicated.
(G) Immunofluorescence images of PC3 cells overexpressing GFP-LARP4A, GFP-LARP4B, or empty GFP plasmid. Scale bars, 100μm.
(H and I) Cell area (H) and circularity (I) of PC3 cells overexpressing LARP4A and LARP4B versus an empty plasmid control. Lines in violin plots depict the mean, n = 600–1000 GFP+ cells from 3 independent experiments.
(J) Immunofluorescence images of MG63 cells overexpressing GFP-LARP4A, GFP-LARP4B, or empty GFP plasmid. Scale bars, 100μm.
(K and L) Cell area (K) and circularity (L) of MG63 cells overexpressing LARP4A and LARP4B versus an empty plasmid control. Lines in violin plots depict the mean, n = 600–1000 GFP+ cells from 3 independent experiments. p values are indicated. See also Figure S10.
Figure 9.
LARP4A and LARP4B regulate cell migration and invasion
(A and B) Wound closure assays in PC3 (A) and MG63 (B) cells over 24-48h either non-transfected (NTC) or transfected with non-targeting control (siCon) and siRNAs targeting LARP4A (si4A1, si4A2) or LARP4B (si4B1, si4B2) as indicated. Blue lines depict the area of the wound at t = 0h and black/white image shows the mask used for wound area analysis. Scale bars, 200μm.
(C and D) Quantification of would area closure by PC3 (C) and MG63 (D) cells over the experimental time course shown in (A and B). The data indicate the mean ± SEM, n = 12–22 regions of interest from 3 independent experiments. p values are indicated.
(E and F) Individual migration tracks of 20 randomly selected PC3 (E) and MG63 (F) control (siControl), siLARP4A, or siLARP4B knockdown cell tracks during a 16h time lapse experiment.
(G and H) Migration analysis (velocity, accumulated distance and Euclidean distance) following the quantification of cell tracks from PC3 (G) and MG63 (H) cell either non-transfected (NTC, siCon) or transfected with siRNAs targeting LARP4A (si4A1, si4A2) or LARP4B (si4B1, si4B2). The data represent the mean ± SEM, n = 65–180 cells from 3 independent experiments. p values are indicated.
(I and J) Brightfield images of transwells stained with crystal violet following a 24h migration assay of PC3 (I) and MG63 (J) control (siControl), siLARP4A (4A1, 4A2), or siLARP4B (4B1, 4B2) knockdown cells. Scale bars, 200μm.
(K and L) Quantification of migrated PC3 (K) and MG63 (L) cells over 24h of either non-transfected cells (NTC, siCon) or cells transfected with siRNAs targeting LARP4A (si4A1, si4A2) or LARP4B (si4B1, si4B2). The data represent the mean ± SEM, n = 15 from 3 independent experiments. p values are indicated. See also Figure S11 and Videos S1, S2, S3, S4, S5, S6, S7, S8, S9, S10, S11, and S12.
Taken together, our results show that both LARP4 paralogs induce morphological changes that are associated with transformed cells and promote cancer cell migration, which may underpin their function in promoting the metastatic potential of cancer cells in vivo.
Discussion
The La-related proteins are emerging as key RBPs with essential roles in RNA biology, but their contributions to disease, including cancer, are not well understood. For the paralogs LARP4A and LARP4B, their roles in cancer progression are only just beginning to be recognised.35 In this work, we demonstrate that LARP4A/4B depletion inhibits osteosarcoma and prostate cancer formation and growth in vivo, advocating for pro-tumorigenic roles of these proteins in these cancers. Additionally, we performed a long overdue systematic comparative analysis of these paralogs, identifying similarities and differences between these two proteins and uncovering potential mechanisms underlying their functions.
The most striking observation was the marked inhibition of tumor growth in xenografts lacking LARP4A or LARP4B in human cancer lines of different origins, mesenchymal and epithelial, as represented by osteosarcoma (MNNG/HOS; MG63) and prostate cancer (PC3) cells, respectively. This is consistent with the high endogenous expression of both LARP4 paralogs in osteosarcoma and prostate cancer. Silencing of either LARP4A or LARP4B inhibited cell viability in vitro and the mitotic indices of both prostate cancer and osteosarcoma cell xenografts in vivo, suggesting that both LARP4 proteins are positive regulators of cell proliferation. The well-established role of LARP4A and LARP4B in sustaining translation rates23,24,25 plausibly impacts on cell proliferation, and this was further corroborated here by our RNAseq analysis reporting elevated mRNA levels of negative regulators of translation and promoters of polyA tail shortening in LARP4A/B depleted cells (AGO2/4, CPEB2, BTG2, TNRC6C, and ZFP36; Figure S12). However, additional cellular mechanisms are likely to play a role. While apoptotic rates were not affected by either LARP4A or LARP4B knockdown, our multiparametric analysis dissecting the consequences of each LARP4 protein knockdown on individual cell cycle stages revealed a novel role for LARP4B in promoting cell cycle progression, specifically affecting entry into mitosis of both cancer cell lines. This was validated by our unbiased genome-wide analyses, revealing that LARP4B regulates a series of targets with GO terms enriched in the regulation of mitosis, spindle organization, and cell cycle phase transition. Cross-examination of differentially expressed genes and candidate LARP4B mRNA targets from a previous PAR-CLIP study in HEK293 cells25 suggests that LARP4B may directly modulate a subset of its RNA targets at the steady-state level to promote mitotic progression. Specific targets that are co-ordinately regulated by LARP4B in a direction that is consistent with mitotic entry and general cell cycle progression include several cyclins and other key cell cycle regulators, namely AURKA/B, E2F1, CCNB1, CCNE2, CDC7, RCF3, MKI67, BRCA2, CDK1/2, and CDKN1A/B/C. Our evidence that LARP4B upregulates some of these transcripts and enhances the translation of AURKB, E2F1, CCNB1, and CCNE2 mRNAs establishes these as potential bona fide LARP4B targets with tumor promoting functions. Indeed, for many cancers, including prostate cancer and osteosarcoma, altered cell cycle control represents a hallmark of tumor progression2 and cyclins and cell cycle kinases are well-established targets for therapy.58,59 For example, AURKB is a mitotic checkpoint kinase dysregulated in many tumors, being linked to tumor cell invasion, metastasis, and drug resistance,60 but is also overexpressed in osteosarcoma61 and its expression directly correlates with Gleason grade in prostate cancer.62 The bidirectional transcriptional regulation of CCNE2 and E2F is critical for cell cycle progression. Moreover, E2F binds to the retinoblastoma (RB1) tumor suppressor protein, which is mutated in many cancers and, together with TP53, is an established driver of prostate cancer and represents a major germline mutation in osteosarcoma.63,64,65 Although caution must be exercised when analysing datasets from different cell lines in different contexts, our data point to a subset of regulated cell cycle, cell proliferation, and oncogenesis-related mRNAs that potentially represent a novel LARP4B RNA regulon,66,67 mediating LARP4B’s effects in driving cancer progression, and importantly, that this appears to be a non-overlapping function with LARP4A.
It has been suggested recently that LARP4A affects cell proliferation rates by regulating levels of proteins essential to mitochondrial function.57 Through its recruitment to mitochondria by the scaffolding factor dAKAP1, LARP4A acts as a general translation factor for the local synthesis of metabolic enzymes that operate the TCA cycle and promote mitochondrial electron transport. LARP4 can also bind to nuclear-encoded mitochondrial mRNAs, regulating their translation.31,57 LARP4A and LARP4B have always been described as cytoplasmic proteins, however, our demonstration that both co-localize to the mitochondria in prostate cancer and osteosarcoma cells supports an essential role for these paralogs in mitochondrial function. To our knowledge, this is the first evidence of mitochondrial localization of LARP4B and is consistent with the identification of mitochondria-associated genes in the LARP4B interactome.25 Together with our bulk RNAseq analysis showing that knockdown of either LARP4A or LARP4B resulted in the overexpression of genes enriched for the response to oxidative stress pathways and glycolysis (Figures 6E and S13), our data further endorse the importance of LARP4 proteins in mitochondrial-elated activities. While the functional implications of these observations remain to be investigated, we suggest the possibility that crosstalk between energy production and LARP4 protein function at mitochondrial membranes may be consistent with the capacities of LARP4A and LARP4B in energetically demanding processes such as cellular proliferation, migration, and cancer development.68,69,70 In addition to subcellular localization to mitochondria, we demonstrated additional nuclear localization in vitro and significantly, in primary murine bone tumors and metastatic lesions in vivo, as well as in human TMAs of osteosarcoma and prostate adenocarcinoma, which hints at a more complex functional profile than previously anticipated. Whereas LARP4A was found to shuttle with PABPC1 to the nuclei during accelerated mRNA decay,71 this is the first evidence of a nuclear localization for LARP4B. This might suggest that the roles of LARP4A and LARP4B in RNA regulation extend to nuclear transcripts and/or involve coordinated cytoplasmic/nuclear activities, and notably, LARP4A and LARP4B have both been identified as nuclear RNA interactors in a global capture study of the human cell nucleus.72 Investigating further the functional significance of such subcellular localizations, and how these contribute to malignant cell growth and migration would be of prime importance.
Knockdown of LARP4A and LARP4B in osteosarcoma cells markedly reduced their ability to seed and expand in lung tissue in a xenograft metastasis model. This is consistent with the high levels of LARP4 proteins in lung nodules formed in a spontaneous osteosarcoma metastasis model, suggesting for the first time, significant pro-invasion and pro-metastatic roles for these proteins in osteosarcoma, although this remains to be determined for prostate cancer. Whether LARP4 proteins regulate extravasation or have specific interactions with endothelial cells is not yet known; however, our unbiased transcriptomic analysis of cells lacking either LARP4A or LARP4B revealed targets that modulate cell junction assembly, cell migration, and ECM organization (Figure S13). Indeed, our in vitro studies demonstrate LARP4A and LARP4B’s regulation of cell morphology, wound healing, and migratory capacity, all characteristic features of metastatic cells. LARP4A is a known regulator of cell morphology and migration, possibly via its interaction with the mechanosensory filamin A (FLNA).73 This function is likely to be context-dependent,37,38 whereas to our knowledge, this is a new cellular function assigned to LARP4B. Interestingly, the effects of LARP4A and LARP4B, particularly for epithelial-derived PC3 cells, appear to be independent of any changes in genes associated with epithelial-mesenchymal transition (EMT) which is a widely accepted mechanism of carcinoma cell metastasis. Moreover, the emerging role of mitochondrial activity in regulating cancer cell migration and metastasis74,75,76 provides a potential novel regulatory link between LARP4A/B mitochondrial localization and their biological effects which should be explored in further research.
In summary, we provide the first comparative in vivo study of the functions and carcinogenic signatures of LARP4A and LARP4B in malignancy development and metastasis in specific cancer models and found that their pro-tumorigenic capacities in this context are largely shared. Uncovering targets and mechanisms of regulation for LARP4 proteins indicate that although some cellular functions may have been preserved throughout evolution, mechanistic differences do also exist. Indeed, there were cell- and gene-specific alterations in the LARP4A- and LARP4B-depleted cancer cell transcriptomes, of which only a small proportion were shared between LARP4A and LARP4B. The increasing investigations of LARP4A and LARP4B in different cancer models indicate complex cell-, tissue- and context-dependent functions of LARP4 proteins in tumor progression.35 We suggest that LARP4A and LARPB be added to the growing list of RBPs that play important roles in osteosarcoma77 and prostate cancer,78 and it will be important to address the intricacies of the underlying molecular mechanisms, cell/tissue-specific RNA targets, and regulatory networks behind LARP4A and LARP4B activities in further functional studies. Nevertheless, our combined in vitro and in vivo findings provide unambiguous evidence of the pro-migratory, growth-promoting and oncogenic potentials of LARP4A and LARP4B osteosarcoma and prostate cancer that may be exploited for biotechnological and therapeutic applications.
Limitations of the study
Although this study revealed a role for LARP4B in the cell cycle regulation and a possible role of LARP4A in cell migration, the mechanistic details and potential RNA targets were not elucidated. Subcellular localization of LARP4A and LARP4B in the nucleus and mitochondria were unveiled in both cancer cell lines and primary human tumor tissue, however, the functional significance of such localization was not explored. We also demonstrated high expression of LARP4A and 4B in a model of osteosarcoma metastasis, however, the functional roles of both paralogs remain to be elucidated using functional osteosarcoma and prostate cancer xenograft studies. Animal studies were not all performed on mixed sex backgrounds therefore the influence of sex/gender on the results of the in vivo studies could not be assessed. Knockdowns were performed using siRNAs that were stable for at least 7days, thus our findings likely underestimate the biological effects of LARP4A and LARP4B on tumor growth. None of these limitations diminish the impact or novelty of the research.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-LARP4A | Abcam | ab241489; RRID: AB_3094705 |
| Rabbit anti-LARP4A | Merck | HPA039306; RRID: AB_3094712 |
| Rabbit anti-LARP4B | Abcam | ab197085; RRID: AB_3094706 |
| Rabbit anti-LARP4B | Merck | HPA036566; RRID: AB_10673624 |
| Rabbit anti-GAPDH | Cell Signalling Technology | 5174; RRID: AB_10622025 |
| Rabbit α-tubulin | Cell Signalling Technology | 2144; RRID: AB_2210548 |
| Mouse anti-E-cadherin | Abcam | ab76055; RRID: AB_1310159 |
| Rabbit anti-Vimentin | Cell Signalling Technology | 5741; RRID: AB_10695459 |
| Mouse anti-TOM20 | Santa Cruz Biotechnology | sc17764; RRID: AB_628381 |
| Mouse Lamin A/C | Santa Cruz Biotechnology | sc7292; RRID: AB_627875 |
| Rabbit anti-E2F1 | Cell Signalling Technology | 3742; RRID: AB_2096936 |
| Rabbit anti-Aurora B | Abcam | ab2254; RRID: AB_302923 |
| Rabbit anti-Cyclin B1 | Cell Signalling Technology | 4138; RRID: AB_2072132 |
| Rabbit anti-Cyclin E2 | Cell Signalling Technology | 4132; RRID: AB_2071197 |
| Goat anti-rabbit HRP-conjugated IgG | Jackson Immunoresearch | 111-035-003; RRID: AB_2313567 |
| Horse anti-mouse HRP-conjugated IgG | Cell Signalling Technology | 7076; RRID: AB_330924 |
| Rabbit anti-Ki67 | Abcam | ab15580; RRID: AB_443209 |
| Chicken anti-GFP | Abcam | ab13970; RRID: AB_300798 |
| Rabbit anti-Cleaved Caspase 3 | Cell Signalling Technology | 9661; RRID: AB_2341188 |
| Rabbit anti-pHH3 | Cell Signalling Technology | 9701; RRID: AB_331535 |
| Goat anti-Rabbit AlexaFluor 488 | Invitrogen | A11034; RRID: AB_2576217 |
| Goat anti-Rabbit AlexaFluor 546 | Invitrogen | A11035; RRID: AB_2534093 |
| Goat anti-Chick AlexaFluor 647 | Abcam | ab150171; RRID: AB_2921318 |
| Goat anti-mouse AlexaFluor 647 | Invitrogen | A21236; RRID: AB_2535805 |
| Bacterial and virus strains | ||
| XL-10 Gold Ultracompetent cells | Agilent | 200314 |
| Biological samples | ||
| Prostate cancer tissue array | Insight Bio | PR481 |
| Bone and cartilage tumour tissue array | Insight Bio | OS804d |
| Normal bone tissue array | Insight Bio | BO244g |
| Chemicals, peptides, and recombinant proteins | ||
| Lipofectamine 2000 | Invitrogen | 11668027 |
| Lipofectamine RNAiMAX | Invitrogen | 13778100 |
| OptiMEM serum-free medium | Gibco | 31985062 |
| mmLVRT reverse transcriptase | Promega | M1701 |
| Brilliant II SYBR Green QPCR Master Mix | Agilent | 600828 |
| Laemmli buffer | Bio-Rad | 1610747 |
| 2-mercaptoethanol | Merck | M3701 |
| Clarity ECL substrates | Bio-Rad | 1705060 |
| Fluoroshield with DAPI | Abcam | Ab104139 |
| DAB subtrate kit | VectorLabs | SK-4100 |
| ProLong Gold Antifade Mountant | Invitrogen | P36930 |
| Phalloidin-TRITC | Merck | P1951 |
| Annexin V AlexaFluore 350 conjugate | Invitrogen | A23202 |
| Propridium Iodide | Invitrogen | P3566 |
| Matrigel | Corning | 354277 |
| Xenolight D-luciferin substrate | PerkinElmer | 122799 |
| CellTrackerTM Green CMFDA Dye | Invitrogen | C2925 |
| Critical commercial assays | ||
| RNeasy Mini kit | Qiagen | 74004 |
| Bicinchoninic acid standard curve assay | Pierce | 23225 |
| Standard cell fractionation kit | Abcam | ab109719 |
| AlexaFluor 647 Click-iT EdU Proliferation kit | Invitrogen | C10340 |
| CellTitre 96 AQueous One Solution MTS | Promega | G3582 |
| Deposited data | ||
| Raw bulk RNAseq data | This paper | GEO: GSE222467 |
| Experimental models: Cell lines | ||
| Human: MG63 osteosarcoma | ATCC | CRL-1427 |
| Human: PC3 prostate cancer | Dr. Magali Williamson, King’s College London | CRL-1435 |
| Human: MNNG/HOS osteosarcoma +GFP/Luc reporter | From Tattersall et al.51 | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: CB17/lcr-Prkdcscid/lcrlcoCrl | Charles River Laboratories | N/A |
| Mouse: Balb/c Nude | Charles River Laboratories | N/A |
| Oligonucleotides | ||
| ON-TARGETplus Non-targeting siRNA #2 | Horizon Discovery | D-001810-02 |
| siGENOME Human LARP4 siRNA | Horizon Discovery | D-016523-02 |
| siGENOME Human LARP4 siRNA | Horizon Discovery | D-016523-04 |
| siGENOME Human LARP4B siRNA | Horizon Discovery | D-026388-01 |
| siGENOME Human LARP4B siRNA | Horizon Discovery | D-026388-04 |
| Recombinant DNA | ||
| pCB6-GFP plasmid | Laboratory of Anne Ridley | N/A |
| pCB6-GFP-LARP4A | This paper | N/A |
| pCB6-GFP-LARP4B | This paper | N/A |
| pCMV2-FLAG-LARP4A | From Yang et al.24 | N/A |
| Software and algorithms | ||
| ImageLab | Bio-Rad | N/A |
| QuPath | From Bankhead et al.79 | N/A |
| ImageJ | National Institute of Health | N/A |
| JACoP | From Bolte et al.80 and Schneider et al.81 | N/A |
| Harmony with PhenoLOGIC | PerkinElmer | N/A |
| MetaMorph | Molecular Devices | N/A |
| Chemotaxis and Migration tool | Ibidi | N/A |
| SkyScan NRecon software v.1.6.9 | Bruker | N/A |
| FastQC | From Andrews82 | N/A |
| HISAT2 | From Kim et al.83 | N/A |
| featureCounts | From Liao et al.84 | N/A |
| EdgeR | From Liu et al.85 and Robinson et al.86 | N/A |
| Enrichr | From Chen et al.87 | N/A |
| Prism | GraphPad | N/A |
| Other | ||
| NuPAGE 4-12% Bis-Tris gels | Invitrogen | NP0321BOX |
| MES running buffer | Invitrogen | NP000202 |
| 8μm pore Transwell chambers | Thermo Scientific | 140656 |
Resource availability
Lead contact
Further information and request for resources should be directed to the lead contact Agamemnon Grigoriadis (agi.grigoriadis@kcl.ac.uk).
Materials availability
Any materials generated from this study are available upon request from the corresponding authors/lead contact.
Data and code availability
-
•
All raw bulk RNAseq data are available in GEO under the accession number GSE222467 and are publicly available.
-
•
This paper does not report original code.
-
•
All other additional information and raw data are available from the lead contact upon request.
Experimental model and study participant details
Ethical approvals and animal usage
All procedures complied with the UK Animals (Scientific Procedures) Act 1986 and were reviewed and approved by the local Research Ethics Committee at KCL. Mice used for subcutaneous and intravital experiments were maintained at the King’s College London (KCL) Biological Services Unit (BSU) under a UK Home Office Project License (P3EBB2E64), while mice used in paratibial xenograft models were maintained at The University of Sheffield (TUoS) BSU Unit under a UK Home Office Project License (PP1668508). For subcutaneous and intravital xenografts, 6-week-old immunocompromised male SCID mice (CB17/lcr-Prkdcscid/lcrlcoCrl) were used. For paratibial models, 6-week-old female Balb/c nude mice were used. All mice for in vivo experiments were purchased from Charles River Laboratories International Inc. and were acclimatised for one week at a minimum in their respective units prior to experimental manipulation. KCL and UoS BSU staff undertook daily husbandry procedures during the experimental courses.
Method details
Constructs
DNA fragments of LARP4A and LARP4B cDNA were subcloned previously into the pCB6-GFP plasmid to obtain N-terminal GFP-tagged LARP4 proteins. cDNA of LARP4A was obtained through restriction enzyme digest of the pCMV2-FLAG-LARP4A plasmid, kindly donated by Dr Richard Maraia (NIH).24 LARP4B cDNA was subcloned similarly from a LARP4B-IMAGE clone (Source Bioscience). Both cDNA fragments were separately purified and ligated into the empty pCB6-GFP plasmid, and the resulting pCB6-GFP-LARP4A and pCB6-GFP-LARP4B plasmids were verified by Sanger sequencing (Source Bioscience).
Cell culture
MG63 cells (male osteosarcoma cell line) were purchased from ATCC (CRL-1427), PC3 cells (male prostate cancer cell line) were donated by Dr. Magali Williamson (King’s College London, KCL) and MNNG/HOS cells (female osteosarcoma cell line) previously transduced with a dual GFP/Luciferase reporter51 were kindly donated by Professor Alison Gartland (University of Sheffield). Osteosarcoma cells (MG63, MNNG/HOS) were cultured in a-MEM containing ultra-glutamine (Lonza) and PC3 cells were cultured in RPMI 1640 (Gibco) supplemented with 2mM L-Glutamine (Merck). All media were supplemented with 10% FBS (Lonza) and 1% penicillin/streptomycin (Merck) and cultured at 37°C in a humidified incubator (5% CO2). Cells were cultured at low passage numbers and passaged consistently using Trypsin-EDTA 0.25% (Merck) and tested regularly for mycoplasma contamination (all cells used for experiments were mycoplasma negative at the time of testing).
Cell transfections
To overexpress GFP-tagged LARP4A and LARP4B, cells were plated at high density 24h prior to transfection using Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions. Transient transfections were performed using Lipofectamine and DNA diluted in Opti-MEM serum-free medium (Gibco) at a 3:1 ratio. Liposome/DNA complexes were added to cells cultured in antibiotic-free medium and incubated for a further 24h before being used for experiments. An empty plasmid control (pCB6-GFP-empty) was used as a control for overexpression experiments.
To deplete LARP4A and LARP4B, two siRNAs targeting each gene (Horizon Discovery) were transfected into cells plated at high density 24h prior, alongside a non-transfected control (NTC, lipofectamine and no siRNA) and a non-targeting siRNA control (lipofectamine and siControl). Cells were transfected at a final concentration of 10nM siRNA using Lipofectamine RNAiMAX (Invitrogen) according to manufacturer’s instructions. In short, siRNA oligos and Lipofectamine RNAiMAX were diluted and mixed in Opti-MEM serum-free medium, after which complexes were added to cells in antibiotic-free medium. Transfected cells were then cultured for 72h before harvesting knockdown confirmation and downstream analysis.
RNA extraction and quantitative PCR
Total RNA was extracted using the RNeasy Mini kit (Qiagen) according to manufacturer’s instructions and quantified using a Nanodrop UV-Vis Spectrophotometer (Thermofisher). RNA-to-cDNA synthesis was performed using dNTPs and mmLVRT reverse transcriptase (Promega) according to manufacturer’s protocol and diluted to a standard concentration for mRNA expression analysis. qPCR was performed using Brilliant II SYBR Green (Agilent) and pre-designed primers (Integrated DNA Technologies) on the LightCycler 480 II (Roche). Prior to experiments, primer efficiency tests and melt curve analyses were performed to ensure specificity of amplified products from all primer pairs. For relative mRNA expression analysis, Ct values were calculated for each amplified product (performed in triplicate) and normalised β-actin mRNA, which was previously determined to be a suitable housekeeping gene, then further to the experimental control using the 2-ΔΔCt method.
Subcellular fractionation and Western blotting
Cell pellets were lysed using urea lysis buffer (1.6M urea, 4.8% SDS, 19.5mM sucrose). Total protein content was quantified using a bicinchoninic acid (BCA) standard curve assay (Pierce) and reagent absorbance determined using the ClarioStar plate reader (BMG Labtech) at 562nm wavelength. Samples at a standard protein concentration were diluted in Western blot sample buffer consisting of Laemmli buffer (Bio-Rad) and 5% β-mercaptoethanol (Merck). Subcellular fractionation of cytoplasmic, mitochondrial and nuclear fractions were isolated from cells in 10cm dishes using the Standard Cell Fractionation Kit (Abcam) according to manufacturer’s instructions. Fraction purity was determined using SDS-PAGE using the nuclear marker Lamin A/C, the mitochondrial marker TOM20 and α-tubulin as a cytoplasmic protein.
Equal volumes of Western blot samples were loaded onto NuPAGE 4-12% Bis-Tris gels (Invitrogen) alongside molecular weight protein standards (New England Biosciences) and subject to SDS-PAGE in MES running buffer (Invitrogen). Proteins were transferred onto PVDF membranes using a semi-dry transfer system (Bio-Rad) and blocked with 5% (w/v) skimmed milk powder (Applichem) diluted in TBS-Tween (0.1%) at room temperature for 1h. Primary antibodies were diluted in blocking buffer and incubated on membranes overnight at 4°C. After several TBS-Tween washes, membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies for another hour at room temperature, before the chemiluminescent signal was detected using Clarity ECL substrates (Bio-Rad) on the ChemiDoc Touch imaging system (Bio-Rad). Protein expression was semi-quantified by calculating relative band intensity of target proteins using ImageLab (Bio-Rad) and normalising to those of housekeeping proteins (GAPDH or α-tubulin). Antibodies used: LARP4A (Abcam, ab241489, 1:1000), LARP4B (Abcam, ab197085, 1:2000), GAPDH (Cell Signalling Technology, CST 5174, 1:3000), α-tubulin (CST 2144, 1:3000), E-cadherin (Abcam, ab76055, 1:200), Vimentin (CST 5741, 1:1000), TOM20 (Santa-Cruz, sc-17764, 1:200), Lamin A/C (Santa-Cruz, sc7292, 1:1000), E2F1 (CST 3742, 1:1000), Aurora B (Abcam, ab2254, 1:500), Cyclin B1 (CST, 4138, 1:1000), Cyclin E2 (CST, 4132, 1:1000), Goat anti-rabbit HRP-conjugated IgG (Stratech, 111-035-003, 1:3000), Horse anti-mouse HRP-conjugated IgG (CST, 7076, 1:3000).
Immunohistochemical and immunofluorescent staining of tissues
Prior to tissue processing, all (bone, tumour and lung) tissues were fixed in 4% PFA overnight and calcified tissues were further decalcified with EDTA. Sections of paraffin-embedded tissues or TMAs were de-waxed and rehydrated in xylene and a graded ethanol series followed by antigen retrieval. This was performed in a citrate buffer for soft tissues (lungs, subcutaneous tumours) and TMAs, or Tris-EDTA buffer for murine bone tissue and osteosarcomas. Sections were blocked for 1h at room temperature in blocking buffer (composed of serum, BSA and Tween-20 diluted in PBS) and primary antibodies, also diluted in blocking buffer, were incubated on tissues overnight at 4°C in a humidified slide box. For immunofluorescence stains, incubation with fluorophore-conjugated secondary antibodies was performed for a further 1h at room temperature. Sections were then washed and mounded with coverslips in Fluoroshield with DAPI (Abcam). For immunohistochemical stains, 3% hydrogen peroxide (Merck) was applied to the sections prior to incubation with HRP-conjugated secondary antibodies for 1h at room temperature. Peroxidase substrates were stained using a DAB kit (VectorLabs), followed by Mayer’s Haematoxylin for 5min. Samples were dehydrated through an increasing ethanol series before being mounted with coverslips using DPX (Merck). For histological staining, sections were stained in Mayer’s Haematoxylin for 8-10min following the same rehydrating graded ethanol series. Following rinsing with dH2O and 95% ethanol, an Eosin Y counterstain was employed for a duration between 30 seconds to 2min depending on the tissue type. Samples were rinsed and rehydrated through an ethanol series before being mounted with DPX. Primary antibodies used: anti-LARP4A (Merck, HPA039306, 1:50), anti-LARP4B (Merck, HPA036566, 1:100), anti-Ki67 (Abcam, ab15580, 1:100), anti-GFP (Abcam, ab13970, 1:300), anti-Cleaved Caspase-3 (CST, 9661, 1:200), anti-pHH3(S10) (CST, 9701, 1:400). Secondary antibodies used: Goat anti-Rabbit AlexaFluor 488 (Thermofisher, A11070, 1:200), Goat anti-Rabbit AlexaFluor 546 (Thermofisher, A11035, 1:200), Goat anti-Chick AlexaFluor 647 (Abcam, ab150171, 1:200), Goat anti-Rabbit HRP-conjugated IgG (Stratech, 111-035-003, 1:200). Immunofluorescent lungs from tail vein experiments were imaged using the Axio Observer7 microscope with Apotome 2 (Zeiss). All other histological, immunohistochemical and immunofluorescent tissue stains were imaged using the Hamamatsu 2-HT slide scanner (Hamamatsu). Expression analyses of subcutaneous tumours and TMAs were performed using QuPath platform for bioimage analysis using the cell detection tools (v.3.0)79 and ImageJ (NIH). Relative DAB staining was semiquantified using ImageJ (NIH) by normalising relative intensity to relative intensity of haematoxylin per area of tissue.
Immunocytochemistry
Cells were plated at low density following incubation with DNA or siRNA 24h prior to fixation with 4% paraformaldehyde (PFA, Merck) for 15min at room temperature. Permeabilisation was performed using 0.5% Triton X-100 (Merck) for 5min at 4°C and cells were further blocked in 5% goat serum in PBS for 1h room temperature. Cells were incubated in primary antibodies diluted in blocking buffer overnight at 4°C before fluorescent secondary antibody staining in the dark for 1h at room temperature and DAPI counterstaining for 5min prior to imaging. For confocal microscopy, cells which were previously plated on glass coverslips were mounted on slides using ProLong Gold Antifade Mountant (Invitrogen) and left to cure overnight before microscopy on the SP5 microscope (Leica) equipped with 405 Blue Diode, Argon (450-520mm), DPSS 561 and HeNe 633 lasers. For high-content analysis and other non-confocal microscopy, cells were imaged within their plates in PBS using the Operetta CLX high-content analysis system (PerkinElmer) and the Axio Observer7 with Apotome (Zeiss), respectively. Primary antibodies used: LARP4A (Merck, HPA039306, 1:100), LARP4B (Merck, HPA036566, 1:150), TOM20 (Santa Cruz, sc-17724, 1:200). Secondary antibodies/stains: Phalloidin-TRITC (Merck, P1951, 1:400), Anti-rabbit AlexaFluor 488 (Invitrogen, A11070, 1:1000), Anti-rabbit AlexaFluor 546 (Invitrogen, A11035, 1:500), Anti-Mouse AlexaFluor 647 (Invitrogen, A21236, 1:1000).
Co-localisation analyses
Fixed cells were stained immunocytochemically as described and imaged using confocal microscopy. Pearson’s and Mander’s pixel correlation and co-localisation coefficients were generated using the ImageJ (NIH) “Just Another Co-Localisation Plugin” (JACoP, v.2.0)80,81 and line density profiles of pixel co-occurrence were calculated using the built-in line and plot profile tool.
High content multiparametric cell cycle analysis
For multiparametric cell cycle analysis, cells grown in a 96 well plate were incubated with 10μM EdU from the AlexaFluor 647 Click-iT EdU Proliferation kit (Invitrogen) for 8h before being fixed in 4% PFA for 15min at room temperature. Washed cells were permeabilised in 0.5% Triton X-100 before 50μl Click-iT reaction cocktail (mixed according to manufacturer’s instructions) was added to each well for 30min at room temperature. Cells were blocked in 5% goat serum for 1h then incubated with primary pHH3(S10) antibody (CST, 53348, 1:800) overnight at 4°C. Cells were finally incubated with secondary antibody and Hoechst (1μg/ml) for 1h at room temperature before washing with PBS and analysing on the Operetta CLS with Harmony (PerkinElmer). Cells were imaged at 10X using the following channels: Hoechst 33342, AF546, AF647, EGFP (if applicable) using pipeline building blocks described earlier.52 Complete cells were initially selected based on the Hoechst channel and the ‘Remove Border Objects’ tool, and singlets/doublets were determined using the PhenoLOGIC machine learning software utilising nuclei property parameters (STAR Methods). The intensity properties of all channels were calculated for singlets only and used for analysis. Frequency distributions of Hoechst intensities of all cells were plotted to find the G1 peak and single-cell Hoechst intensities were normalised to the value of this peak to obtain ‘Normalised DNA content’.52 This allowed categorisation of cells into cell cycle stages (sub-G1, G1/S and G2/M) based on their normalised DNA content. Cells could be more specifically categorised into S phase using their Hoechst intensities in combination with their mean EdU incorporation, and into M phase with their pHH3 expression. To perform cell cycle analyses on only GFP-LARP4A/LARP4B expressing singlets in overexpression experiments, an adapted pipeline with an additional building block was used to determine GFP+ cells using GFP intensity properties.
Growth assays
For viability assays, CellTitre 96 AQueous One Solution MTS (Promega) was added to cells plated at low density according to manufacturer’s instructions and incubated in darkness in a humidified incubator for 3h at 37°C, 5% CO2. Absorbances of media containing MTS were then measured at 490nm using the ClarioStar plate reader (BMG LabTech). Each cell condition was measured every 24h over the course of 4d. For cell growth assays, cells were plated in low confluency and left to grow in normal conditions for 72h, after which they were trypsinised and counted using a haemocytometer to determine total cell number.
Apoptosis assays
Cells were collected following transfection incubations and resuspended in Annexin V binding buffer and Annexin V AlexaFluore 350 conjugate (Invitrogen) or Propridium Iodide (PI, Invitrogen) according to instructions. Cell suspensions were incubated in the dark at room temperature for 15min before being analysed by flow cytometry on the LSRFortessa system (BD Biosciences), alongside positive and negative controls, as well as PI- or Annexin V-only controls to calculate compensation. Thresholding was performed using the DAPI (Annexin V) and PE-Texas Red (PI) channels, as well as the EGFP channel for overexpression experiments to determine GFP+ populations. Analysis of apoptotic populations was performed on FlowJo software (BD Biosciences).
Morphological analysis
Cells plated in tissue culture plates pre-coated with Matrigel were fixed and stained using immunocytochemistry as described above. Cells stained with Phalloidin and DAPI were imaged at 10X using TRITC, DAPI and EGFP channels (if applicable, for overexpression experiments). Six regions of interest were taken from each well, with conditions performed in duplicate over three independent experiments. Image files were analysed using ImageJ (NIH). Cell shape parameters, including perimeter, area and circularity, were determined for each cell within the field of view based on phalloidin stains, and calculations for circularity were made (below), producing arbitrary values where 0 indicates a perfect circle and 1 indicates a straight line.38
Migration assays
Determining the influence of LARP4 proteins on cell migration was determined using three independent methods. For scratch assays, cells were plated as a confluent monolayer and serum-starved for 24h prior to the start of the experiment to limit cell division. Wounds were formed in the wells, washed twice and imaged at 4X magnification using phase-contrast time lapse microscopy using the IX81 inverted live-cell light microscope (Olympus) over 24h (PC3 cells) or 48h (MG63 cells). Four regions of interest per scratch were measured over the experiment duration with a frame acquisition of one per 30min. Wound closure over time was quantified using ImageJ, where wound area was quantified at 0h (t=0), and several other time points until 24 or 48h. Wound closure was finally calculated as the percentage change in wound area over time. For single cell migration experiments, cells were plated at low density prior to the start of the experiment in phenol red-free medium. Time-lapse video imaging was performed over 12-18h, with cells being imaged using phase-contrast time lapse microscopy using the IX81 inverted live cell light microscope (Olympus) equipped with MetaMorph time lapse software (Molecular Devices). One frame was captured every 5min and compiled into stack files using MetaMorph software. Stacks were imported into ImageJ (NIH) and cells were manually tracked using the ‘Manual Tracking’ plugin. Cells that divided, died or moved out of the image frame during the course of the experiment were not tracked. All stacks were saved as a .gif video file. Individual cell velocity, accumulated and Euclidean distance were calculated using cell coordinates for each frame using the Chemotaxis and Migration Tool (Ibidi). For transwell migration, cells were seeded onto 8μm pore Transwell chambers (Thermo Scientific) containing serum-free medium, which were further placed into wells containing medium supplemented with 10% FBS. The plates were incubated at 37°C, 5% CO2, for 24h before transwells were washed, fixed in 4% PFA, permeabilised in methanol and stained with crystal violet for visualisation of migrated cells. Non-migrated cells were removed using a swab and transwell membranes imaged using an Olympus-LS brightfield Stereo Microscope (Olympus) and crystal violet positive cells that had migrated to the other side of the transwell were quantified using ImageJ.
Xenograft experiments
For subcutaneous models of osteosarcoma and prostate cancer cell xenografts, 1 million viable MNNG/HOS and PC3 cells that had been transfected with control or LARP4A/LARP4B-targeting siRNAs 72h prior (as described above) suspended in a 100μl volume of 1:1 mix of Matrigel:PBS were injected subcutaneously into both flanks of each mouse (2 sites per mouse; 5-8 mice per group, n=15-24). All mice were randomly allocated and experiments were blinded. Mice were monitored for 15d for tumour growth, at which point mice were sacrificed and tumours excised for further analysis. Tumour dimensions were measured using electronic calipers and volume was calculated using the following formula:
For paratibial models of osteosarcoma xenografts, 500,000 viable MNNG/HOS cells previously transfected with targeting/non-targeting siRNAs were injected onto the surface of both tibiae of anaesthetised mice (isoflurane inhalation, 8 per group, n=24). All mice were randomly allocated and experiments were blinded. Luciferase measurements were taken every other day beginning from day 2 (the day after the injection) – mice were anaesthetised (isoflurane inhalation) and subcutaneously injected with Xenolight D-luciferin substrate (PerkinElmer). Mice were imaged after 5min using the IVIS Lumina II (Caliper Life Sciences), images acquired and luciferase luminescence quantified (total photon flux) using IVIS Living Image software (PerkinElmer). Tumours of control mice became palpable at day 9, after which tibial volumes of mice were measured every 2-3d. Two perpendicular measurements across the tibia were taken and total tibial/tumour volume was calculated using the following formula (ML = Largest measurement; MS = Smallest measurement).
After 14d, mice were sacrificed and tibiae and lungs were dissected for further analysis. Tibiae were fixed for 48h in formalin then switched to 70% ethanol before scanning on a SkyScan 1172 microCT machine (Bruker) at 8μm resolution. The X-Ray source functioned at 200μA, 50kV using a 0.5mm aluminium filter with images captured each 0.7° angle and images were lastly reconstructed using the SkyScan NRecon software (Bruker, v.1.6.9).
Intravital models of metastasis
For the short term intravital model of metastasis, 1 million viable CMFDA-labelled MG63 cells previously transfected with targeting or non-targeting siRNAs were injected into the tail veins of mice (6 per group, n=18) in 100μl PBS and sacrificed 24h later. For the longer-term intravital model, 5 million viable MNNG/HOS cells previously transfected with targeting or non-targeting siRNAs were injected into mice tail veins in 100μl PBS and mice were sacrificed after 7d. For both models, murine lungs were dissected and fixed for histology and immunofluorescent staining.
Tissue mRNA expression analyses (TCGA TARGET GTEX)
Normal tissue versus tumour tissue LARP4A and LARP4B gene expression analysis was performed using the University of California Santa Cruz (UCSC) Xena browser.88,89,90 mRNA expression data from The Cancer Genome Atlas (TCGA) Therapeutically Applicable Research to Generate Effective Treatments (TARGET) and Genotype Tissue Expression Project (GTEx) study were used. mRNA expression values of LARP4A and LARP4B were available in normal and tumour tissue across 23 tissue types. Differences between means of normal versus tissue mRNA expression were analysed using a Welch’s t-test in GraphPad Prism (v.9.1.1).
Bulk RNAseq
RNA was extracted according to the procedure described above from cells previously transfected with siControl siRNA or two siRNAs targeting LARP4A or LARP4B (two independent replicates per condition). High-quality RNA samples, from two independent biological replicates per condition, were submitted to Genewiz (Bahnofstrasse), which further confirmed high RNA integrity using a BioAnalyzer 2100 (Agilent). Following poly(A) selection and library preparation, mRNA was sequenced using the NovaSeq 6000 system (Illumina; paired end, 150bp). FastQC82 confirmed high per base sequence quality and high per sequence quality scores for each individual fastq file and assessed duplicate reads. HISAT283 then mapped reads to the human reference genome (Hg38) and individual count files were generated using ‘featureCounts’.84 EdgeR85,86 was finally used for gene normalisation and differential expression testing between groups. The following were group comparisons used: MG63 siLARP4A or siLARP4B vs siControl; PC3 siLARP4A or siLARP4B vs siControl. Differentially expressed genes (DEGs) were determined using the following standard parameters: false discovery rate (FDR) ≤ 0.05, logFC value of >0.5 (upregulated vs siControl) or <0.5 (downregulated vs siControl). Enrichr87,91,92 was used for gene enrichment analysis using the Gene Ontology (GO) database.
Quantification and statistical analysis
With the exception of RNAseq analysis, which is described in detail above, all statistical analyses were accomplished using GraphPad prism (v.9.1.1). Testing for statistically significant differences in means between two groups was performed using Welch’s t-tests (non-paired), where testing for differences between more than two groups was performed using one-way Analyses of Variances (ANOVA) followed up with Tukey’s multiple comparisons post-hoc tests. Statistical significance was defined using the standard threshold of p<0.05. All experiments were performed using a minimum of three independent biological replicates unless otherwise stated, with between two to four technical replicates per sample. Significance values are either denoted numerically or with asterisks where false discovery rate (FDR) or p value are: ∗ p<0.05; ∗∗ p<0.01, ∗∗∗ p<0.001, ∗∗∗∗ p<0.0001.
Acknowledgments
The authors thank Prof. Peter Parker (Francis Crick Institute, UK), and Dr. Eric Rahrmann (University of Cambridge, UK) for insightful discussion and critical reading of the article. We thank Drs. Richard Maraia and Sandy Mattijssen (NIH, USA) for the gift of the pCMV2-FLAG-LARP4A plasmid and useful discussions. We are grateful to Dr. Isabel Cruz-Gallardo and Jiazhen Shen for cloning pCB6-GFP-LARP4A and LARP4B plasmids. We thank Dr. Matthias Krause (King’s College London) for providing the IX81 microscope set up for the migration experiments, Prof. Anita Grigoriadis (King’s College London) for help with the RNAseq experiment, Dr. Magali Williamson (King’s College London) for providing us with the PC3 cell line, Prof. Tony Ng (King’s College London) for enabling the in vivo studies, and Ms Dhivya Chandrasekaran (King’s College London) for technical help with the xenograft experiments. We thank the BRC Flow Cytometry Core and Biological Services Unit at King’s College London.
This work was supported by a grant from the UK Medical Research Council (MR/N013700/1) awarded to J. Coleman as part of a Doctoral Training Partnership scholarship at King’s College London.
Author contributions
M.R.C., A.J.R., and A.E.G. conceived the study. J.C., M.R.C and A.E.G. planned and designed experiments. J.C. L.T., L.K., H.Y., S.R.H., P.J., A.J.C., C.C., M.R.C., and A.E.G., performed experiments. V.Y. generated, analyzed, and provided expert consultation on bulk RNAseq analysis. L.T. and A.G. performed the paratibial xenografts and subsequent analysis. M.R.C. and A.E.G. supervised the study. J.C., M.R.C., and A.E.G. wrote and edited the article.
Declaration of interests
The authors declare that they have no competing interests.
Published: February 24, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109288.
Contributor Information
Maria R. Conte, Email: sasi.conte@kcl.ac.uk.
Agamemnon E. Grigoriadis, Email: agi.grigoriadis@kcl.ac.uk.
Supplemental information
References
- 1.Bailey M.H., Tokheim C., Porta-Pardo E., Sengupta S., Bertrand D., Weerasinghe A., Colaprico A., Wendl M.C., Kim J., Reardon B., et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell. 2018;173:371–385.e18. doi: 10.1016/j.cell.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 4.Sharp P.A. The Centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Müller-McNicoll M., Neugebauer K.M. How cells get the message: dynamic assembly and function of mRNA–protein complexes. Nat. Rev. Genet. 2013;14:275–287. doi: 10.1038/nrg3434. [DOI] [PubMed] [Google Scholar]
- 6.Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., Davey N.E., Humphreys D.T., Preiss T., Steinmetz L.M., et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 7.Van Nostrand E.L., Freese P., Pratt G.A., Wang X., Wei X., Xiao R., Blue S.M., Chen J.-Y., Cody N.A.L., Dominguez D., et al. A large-scale binding and functional map of human RNA-binding proteins. Nature. 2020;583:711–719. doi: 10.1038/s41586-020-2077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi P.S., Thomas-Tikhonenko A. RNA-binding proteins of COSMIC importance in cancer. J. Clin. Invest. 2021;131 doi: 10.1172/JCI151627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lukong K.E., Chang K.W., Khandjian E.W., Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Kang D., Lee Y., Lee J.-S. RNA-Binding Proteins in Cancer: Functional and Therapeutic Perspectives. Cancers. 2020;12:2699. doi: 10.3390/cancers12092699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maraia R.J., Mattijssen S., Cruz-Gallardo I., Conte M.R. The La and related RNA-binding proteins (LARPs): structures, functions, and evolving perspectives. WIREs RNA. 2017;8 doi: 10.1002/wrna.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bousquet-Antonelli C., Deragon J.M. A comprehensive analysis of the La-motif protein superfamily. RNA. 2009;15:750–764. doi: 10.1261/rna.1478709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dock-Bregeon A.C., Lewis K.A., Conte M.R. The La-related proteins: structures and interactions of a versatile superfamily of RNA-binding proteins. RNA Biol. 2021;18:178–193. doi: 10.1080/15476286.2019.1695712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lizarrondo J., Dock-Bregeon A.-C., Martino L., Conte M.R. Structural dynamics in the La-module of La-related proteins. RNA Biol. 2021;18:194–206. doi: 10.1080/15476286.2020.1733799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martino L., Pennell S., Kelly G., Busi B., Brown P., Atkinson R.A., Salisbury N.J.H., Ooi Z.H., See K.W., Smerdon S.J., et al. Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module. Nucleic Acids Res. 2015;43:645–660. doi: 10.1093/nar/gku1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotik-Kogan O., Valentine E.R., Sanfelice D., Conte M.R., Curry S. Structural analysis reveals conformational plasticity in the recognition of RNA 3' ends by the human La protein. Structure. 2008;16:852–862. doi: 10.1016/j.str.2008.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alfano C., Sanfelice D., Babon J., Kelly G., Jacks A., Curry S., Conte M.R. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat. Struct. Mol. Biol. 2004;11:323–329. doi: 10.1038/nsmb747. [DOI] [PubMed] [Google Scholar]
- 18.Uchikawa E., Natchiar K.S., Han X., Proux F., Roblin P., Zhang E., Durand A., Klaholz B.P., Dock-Bregeon A.C. Structural insight into the mechanism of stabilization of the 7SK small nuclear RNA by LARP7. Nucleic Acids Res. 2015;43:3373–3388. doi: 10.1093/nar/gkv173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozlov G., Mattijssen S., Jiang J., Nyandwi S., Sprules T., Iben J.R., Coon S.L., Gaidamakov S., Noronha A.M., Wilds C.J., et al. Structural basis of 3'-end poly(A) RNA recognition by LARP1. Nucleic Acids Res. 2022;50:9534–9547. doi: 10.1093/nar/gkac696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cruz-Gallardo I., Martino L., Kelly G., Atkinson R.A., Trotta R., De Tito S., Coleman P., Ahdash Z., Gu Y., Bui T.T.T., Conte M.R. LARP4A recognizes polyA RNA via a novel binding mechanism mediated by disordered regions and involving the PAM2w motif, revealing interplay between PABP, LARP4A and mRNA. Nucleic Acids Res. 2019;47:4272–4291. doi: 10.1093/nar/gkz144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stavraka C., Blagden S. The La-Related Proteins, a Family with Connections to Cancer. Biomolecules. 2015;5:2701–2722. doi: 10.3390/biom5042701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grimm C., Pelz J.P., Schneider C., Schäffler K., Fischer U. Crystal Structure of a Variant PAM2 Motif of LARP4B Bound to the MLLE Domain of PABPC1. Biomolecules. 2020;10 doi: 10.3390/biom10060872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schäffler K., Schulz K., Hirmer A., Wiesner J., Grimm M., Sickmann A., Fischer U. A stimulatory role for the La-related protein 4B in translation. RNA. 2010;16:1488–1499. doi: 10.1261/rna.2146910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang R., Gaidamakov S.A., Xie J., Lee J., Martino L., Kozlov G., Crawford A.K., Russo A.N., Conte M.R., Gehring K., Maraia R.J. La-Related Protein 4 Binds Poly(A), Interacts with the Poly(A)-Binding Protein MLLE Domain via a Variant PAM2w Motif, and Can Promote mRNA Stability. Mol. Cell Biol. 2011;31:542–556. doi: 10.1128/mcb.01162-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Küspert M., Murakawa Y., Schäffler K., Vanselow J.T., Wolf E., Juranek S., Schlosser A., Landthaler M., Fischer U. LARP4B is an AU-rich sequence associated factor that promotes mRNA accumulation and translation. RNA. 2015;21:1294–1305. doi: 10.1261/rna.051441.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranjan A., Mattijssen S., Cruz-Gallardo I., Pitman L.F.P.F., Coleman J.C., Conte M.R., Maraia R.J. A conserved region of LARP4 that binds RACK1 promotes ARE-mRNA stabilization and translation. SSRN. 2023 doi: 10.2139/ssrn.4587011. Preprint at. [DOI] [Google Scholar]
- 27.Mattijssen S., Arimbasseri A.G., Iben J.R., Gaidamakov S., Lee J., Hafner M., Maraia R.J. LARP4 mRNA codon-tRNA match contributes to LARP4 activity for ribosomal protein mRNA poly(A) tail length protection. Elife. 2017;6 doi: 10.7554/elife.28889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattijssen S., Iben J.R., Li T., Coon S.L., Maraia R.J. Single molecule poly(A) tail-seq shows LARP4 opposes deadenylation throughout mRNA lifespan with most impact on short tails. Elife. 2020;9 doi: 10.7554/elife.59186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian Y., Zeng Z., Li X., Wang Y., Chen R., Mattijssen S., Gaidamakov S., Wu Y., Maraia R.J., Peng W., Zhu J. Transcriptome-wide stability analysis uncovers LARP4-mediated NFkappaB1 mRNA stabilization during T cell activation. Nucleic Acids Res. 2020;48:8724–8739. doi: 10.1093/nar/gkaa643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim S., Kim S., Chang H.R., Kim D., Park J., Son N., Park J., Yoon M., Chae G., Kim Y.K., et al. The regulatory impact of RNA-binding proteins on microRNA targeting. Nat. Commun. 2021;12:5057. doi: 10.1038/s41467-021-25078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gabrovsek L., Collins K.B., Aggarwal S., Saunders L.M., Lau H.-T., Suh D., Sancak Y., Trapnell C., Ong S.-E., Smith F.D., Scott J.D. A-kinase-anchoring protein 1 (dAKAP1)-based signaling complexes coordinate local protein synthesis at the mitochondrial surface. J. Biol. Chem. 2020;295:10749–10765. doi: 10.1074/jbc.ra120.013454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Funakoshi M., Tsuda M., Muramatsu K., Hatsuda H., Morishita S., Aigaki T. Overexpression of Larp4B downregulates dMyc and reduces cell and organ sizes in Drosophila. Biochem. Biophys. Res. Commun. 2018;497:762–768. doi: 10.1016/j.bbrc.2018.02.148. [DOI] [PubMed] [Google Scholar]
- 33.Deragon J.M. Distribution, organization an evolutionary history of La and LARPs in eukaryotes. RNA Biol. 2021;18:159–167. doi: 10.1080/15476286.2020.1739930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merret R., Martino L., Bousquet-Antonelli C., Fneich S., Descombin J., Billey É., Conte M.R., Deragon J.-M. The association of a La module with the PABP-interacting motif PAM2 is a recurrent evolutionary process that led to the neofunctionalization of La-related proteins. RNA. 2013;19:36–50. doi: 10.1261/rna.035469.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coleman J.C., Hallett S.R., Grigoriadis A.E., Conte M.R. LARP4A and LARP4B in cancer: The new kids on the block. Int. J. Biochem. Cell Biol. 2023;161 doi: 10.1016/j.biocel.2023.106441. [DOI] [PubMed] [Google Scholar]
- 36.Bai S.W., Herrera-Abreu M.T., Rohn J.L., Racine V., Tajadura V., Suryavanshi N., Bechtel S., Wiemann S., Baum B., Ridley A.J. Identification and characterization of a set of conserved and new regulators of cytoskeletal organization, cell morphology and migration. BMC Biol. 2011;9:54. doi: 10.1186/1741-7007-9-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egiz M., Usui T., Ishibashi M., Zhang X., Shigeta S., Toyoshima M., Kitatani K., Yaegashi N. La-Related Protein 4 as a Suppressor for Motility of Ovarian Cancer Cells. Tohoku J. Exp. Med. 2019;247:59–67. doi: 10.1620/tjem.247.59. [DOI] [PubMed] [Google Scholar]
- 38.Seetharaman S., Flemyng E., Shen J., Conte M.R., Ridley A.J. The RNA-binding protein LARP4 regulates cancer cell migration and invasion. Cytoskeleton. 2016;73:680–690. doi: 10.1002/cm.21336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koso H., Takeda H., Yew C.C.K., Ward J.M., Nariai N., Ueno K., Nagasaki M., Watanabe S., Rust A.G., Adams D.J., et al. Transposon mutagenesis identifies genes that transform neural stem cells into glioma-initiating cells. Proc. Natl. Acad. Sci. USA. 2012;109:E2998–E3007. doi: 10.1073/pnas.1215899109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koso H., Yi H., Sheridan P., Miyano S., Ino Y., Todo T., Watanabe S. Identification of RNA-Binding Protein LARP4B as a Tumor Suppressor in Glioma. Cancer Res. 2016;76:2254–2264. doi: 10.1158/0008-5472.CAN-15-2308. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y., Peng L., Hu T., Wan Y., Ren Y., Zhang J., Wang X., Zhou Y., Yuan W., Wang Q., et al. La-related protein 4B maintains murine MLL-AF9 leukemia stem cell self-renewal by regulating cell cycle progression. Exp. Hematol. 2015;43:309–318.e2. doi: 10.1016/j.exphem.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 42.Li Y., Jiao Y., Li Y., Liu Y. Expression of La Ribonucleoprotein Domain Family Member 4B (LARP4B) in Liver Cancer and Their Clinical and Prognostic Significance. Dis. Markers. 2019;2019:1569049. doi: 10.1155/2019/1569049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeda H., Wei Z., Koso H., Rust A.G., Yew C.C.K., Mann M.B., Ward J.M., Adams D.J., Copeland N.G., Jenkins N.A. Transposon mutagenesis identifies genes and evolutionary forces driving gastrointestinal tract tumor progression. Nat. Genet. 2015;47:142–150. doi: 10.1038/ng.3175. [DOI] [PubMed] [Google Scholar]
- 44.Bard-Chapeau E.A., Nguyen A.T., Rust A.G., Sayadi A., Lee P., Chua B.Q., New L.S., de Jong J., Ward J.M., Chin C.K., et al. Transposon mutagenesis identifies genes driving hepatocellular carcinoma in a chronic hepatitis B mouse model. Nat. Genet. 2014;46:24–32. doi: 10.1038/ng.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rahrmann E.P., Watson A.L., Keng V.W., Choi K., Moriarity B.S., Beckmann D.A., Wolf N.K., Sarver A., Collins M.H., Moertel C.L., et al. Forward genetic screen for malignant peripheral nerve sheath tumor formation identifies new genes and pathways driving tumorigenesis. Nat. Genet. 2013;45:756–766. doi: 10.1038/ng.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu X., Northcott P.A., Dubuc A., Dupuy A.J., Shih D.J.H., Witt H., Croul S., Bouffet E., Fults D.W., Eberhart C.G., et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 2012;482:529–533. doi: 10.1038/nature10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu Z., Yau C., Ahmed A.A. A pan-cancer genome-wide analysis reveals tumour dependencies by induction of nonsense-mediated decay. Nat. Commun. 2017;8 doi: 10.1038/ncomms15943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grigoriadis A.E., Schellander K., Wang Z.Q., Wagner E.F. Osteoblasts are target cells for transformation in c-fos transgenic mice. J. Cell Biol. 1993;122:685–701. doi: 10.1083/jcb.122.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weekes D., Kashima T.G., Zandueta C., Perurena N., Thomas D.P., Sunters A., Vuillier C., Bozec A., El-Emir E., Miletich I., et al. Regulation of osteosarcoma cell lung metastasis by the c-Fos/AP-1 target FGFR1. Oncogene. 2016;35:2852–2861. doi: 10.1038/onc.2015.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeng W.Z.D., Glicksberg B.S., Li Y., Chen B. Selecting precise reference normal tissue samples for cancer research using a deep learning approach. BMC Med. Genomics. 2019;12 doi: 10.1186/s12920-018-0463-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tattersall L., Shah K.M., Lath D.L., Singh A., Down J.M., De Marchi E., Williamson A., Di Virgilio F., Heymann D., Adinolfi E., et al. The P2RX7B splice variant modulates osteosarcoma cell behaviour and metastatic properties. J. Bone Oncol. 2021;31 doi: 10.1016/j.jbo.2021.100398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Massey A.J. Multiparametric Cell Cycle Analysis Using the Operetta High-Content Imager and Harmony Software with PhenoLOGIC. PLoS One. 2015;10 doi: 10.1371/journal.pone.0134306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang C.-C., Kato H., Shindo M., Masai H. Cdc7 activates replication checkpoint by phosphorylating the Chk1-binding domain of Claspin in human cells. Elife. 2019;8 doi: 10.7554/elife.50796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Denechaud P.D., Fajas L., Giralt A. E2F1, a Novel Regulator of Metabolism. Front. Endocrinol. 2017;8:311. doi: 10.3389/fendo.2017.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uxa S., Castillo-Binder P., Kohler R., Stangner K., Müller G.A., Engeland K. Ki-67 gene expression. Cell Death Differ. 2021;28:3357–3370. doi: 10.1038/s41418-021-00823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Misra S., Sharma S., Agarwal A., Khedkar S.V., Tripathi M.K., Mittal M.K., Chaudhuri G. Cell cycle-dependent regulation of the bi-directional overlapping promoter of human BRCA2/ZAR2 genes in breast cancer cells. Mol. Cancer. 2010;9:50. doi: 10.1186/1476-4598-9-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lewis B.M., Cho C.Y., Her H.-L., Hunter T., Yeo G.W. LARP4 Is an RNA-binding protein that binds nuclear-encoded mitochondrial mRNAs to promote mitochondrial function. RNA. 2024;30:223–239. doi: 10.1261/rna.079799.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ben-Salem S., Venkadakrishnan V.B., Heemers H.V. Novel insights in cell cycle dysregulation during prostate cancer progression. Endocr. Relat. Cancer. 2021;28:R141–R155. doi: 10.1530/ERC-20-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng L., Wang C., Jing J. Cell Cycle Kinases in Osteosarcoma: Potential for Therapeutic Intervention. Curr. Pharm. Des. 2016;22:4830–4834. doi: 10.2174/1381612822666160512151028. [DOI] [PubMed] [Google Scholar]
- 60.Bavetsias V., Linardopoulos S. Aurora Kinase Inhibitors: Current Status and Outlook. Front. Oncol. 2015;5:278. doi: 10.3389/fonc.2015.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu L.B., Jiang J., Zhu X.P., Wang T.F., Chen X.Y., Luo Q.F., Shu Y., Liu Z.L., Huang S.H. Knockdown of Aurora-B inhibits osteosarcoma cell invasion and migration via modulating PI3K/Akt/NF-kappaB signaling pathway. Int. J. Clin. Exp. Pathol. 2014;7:3984–3991. [PMC free article] [PubMed] [Google Scholar]
- 62.Chieffi P., Cozzolino L., Kisslinger A., Libertini S., Staibano S., Mansueto G., De Rosa G., Villacci A., Vitale M., Linardopoulos S., et al. Aurora B expression directly correlates with prostate cancer malignancy and influence prostate cell proliferation. Prostate. 2006;66:326–333. doi: 10.1002/pros.20345. [DOI] [PubMed] [Google Scholar]
- 63.Kansara M., Teng M.W., Smyth M.J., Thomas D.M. Translational biology of osteosarcoma. Nat. Rev. Cancer. 2014;14:722–735. doi: 10.1038/nrc3838. [DOI] [PubMed] [Google Scholar]
- 64.Nyquist M.D., Corella A., Coleman I., De Sarkar N., Kaipainen A., Ha G., Gulati R., Ang L., Chatterjee P., Lucas J., et al. Combined TP53 and RB1 Loss Promotes Prostate Cancer Resistance to a Spectrum of Therapeutics and Confers Vulnerability to Replication Stress. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang W., Zhang K. A transcriptomic signature for prostate cancer relapse prediction identified from the differentially expressed genes between TP53 mutant and wild-type tumors. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-14436-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bisogno L.S., Keene J.D. RNA regulons in cancer and inflammation. Curr. Opin. Genet. Dev. 2018;48:97–103. doi: 10.1016/j.gde.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blackinton J.G., Keene J.D. Post-transcriptional RNA regulons affecting cell cycle and proliferation. Semin. Cell Dev. Biol. 2014;34:44–54. doi: 10.1016/j.semcdb.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Epstein T., Xu L., Gillies R.J., Gatenby R.A. Separation of metabolic supply and demand: aerobic glycolysis as a normal physiological response to fluctuating energetic demands in the membrane. Cancer Metab. 2014;2:7. doi: 10.1186/2049-3002-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shiraishi T., Verdone J.E., Huang J., Kahlert U.D., Hernandez J.R., Torga G., Zarif J.C., Epstein T., Gatenby R., McCartney A., et al. Glycolysis is the primary bioenergetic pathway for cell motility and cytoskeletal remodeling in human prostate and breast cancer cells. Oncotarget. 2015;6:130–143. doi: 10.18632/oncotarget.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zanotelli M.R., Zhang J., Reinhart-King C.A. Mechanoresponsive metabolism in cancer cell migration and metastasis. Cell Metab. 2021;33:1307–1321. doi: 10.1016/j.cmet.2021.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gilbertson S., Federspiel J.D., Hartenian E., Cristea I.M., Glaunsinger B. Changes in mRNA abundance drive shuttling of RNA binding proteins, linking cytoplasmic RNA degradation to transcription. Elife. 2018;7 doi: 10.7554/eLife.37663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Conrad T., Albrecht A.S., de Melo Costa V.R., Sauer S., Meierhofer D., Ørom U.A. Serial interactome capture of the human cell nucleus. Nat. Commun. 2016;7 doi: 10.1038/ncomms11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mao Z., Nakamura F. Interaction of LARP4 to filamin A mechanosensing domain regulates cell migrations. Front. Cell Dev. Biol. 2023;11 doi: 10.3389/fcell.2023.1152109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Denisenko T.V., Gorbunova A.S., Zhivotovsky B. Mitochondrial Involvement in Migration, Invasion and Metastasis. Front. Cell Dev. Biol. 2019;7:355. doi: 10.3389/fcell.2019.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Valcarcel-Jimenez L., Gaude E., Torrano V., Frezza C., Carracedo A. Mitochondrial Metabolism: Yin and Yang for Tumor Progression. Trends Endocrinol. Metab. 2017;28:748–757. doi: 10.1016/j.tem.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yadav T., Gau D., Roy P. Mitochondria-actin cytoskeleton crosstalk in cell migration. J. Cell. Physiol. 2022;237:2387–2403. doi: 10.1002/jcp.30729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang J., Miao X., Wu T., Jia J., Cheng X. Development and Validation of Ten-RNA Binding Protein Signature Predicts Overall Survival in Osteosarcoma. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.751842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gao L., Meng J., Zhang Y., Gu J., Han Z., Wang X., Gao S. Development and validation of a six-RNA binding proteins prognostic signature and candidate drugs for prostate cancer. Genomics. 2020;112:4980–4992. doi: 10.1016/j.ygeno.2020.08.034. [DOI] [PubMed] [Google Scholar]
- 79.Bankhead P., Loughrey M.B., Fernández J.A., Dombrowski Y., McArt D.G., Dunne P.D., McQuaid S., Gray R.T., Murray L.J., Coleman H.G., et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bolte S., Cordelières F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 81.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 83.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 85.Liu R., Holik A.Z., Su S., Jansz N., Chen K., Leong H.S., Blewitt M.E., Asselin-Labat M.L., Smyth G.K., Ritchie M.E. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res. 2015;43:e97. doi: 10.1093/nar/gkv412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Goldman M., Craft B., Hastie M., Repečka K., McDade F., Kamath A., Banerjee A., Luo Y., Rogers D., Brooks A.N. The UCSC Xena platform for public and private cancer genomics data visualization and interpretation. bioRxiv. 2018 doi: 10.1101/326470. Preprint at: [DOI] [Google Scholar]
- 89.Goldman M.J., Craft B., Hastie M., Repečka K., McDade F., Kamath A., Banerjee A., Luo Y., Rogers D., Brooks A.N., et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020;38:675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xie Z., Bailey A., Kuleshov M.V., Clarke D.J.B., Evangelista J.E., Jenkins S.L., Lachmann A., Wojciechowicz M.L., Kropiwnicki E., Jagodnik K.M., et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021;1:e90. doi: 10.1002/cpz1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All raw bulk RNAseq data are available in GEO under the accession number GSE222467 and are publicly available.
-
•
This paper does not report original code.
-
•
All other additional information and raw data are available from the lead contact upon request.