Figure 6.
Identification of LARP4A and LARP4B targets
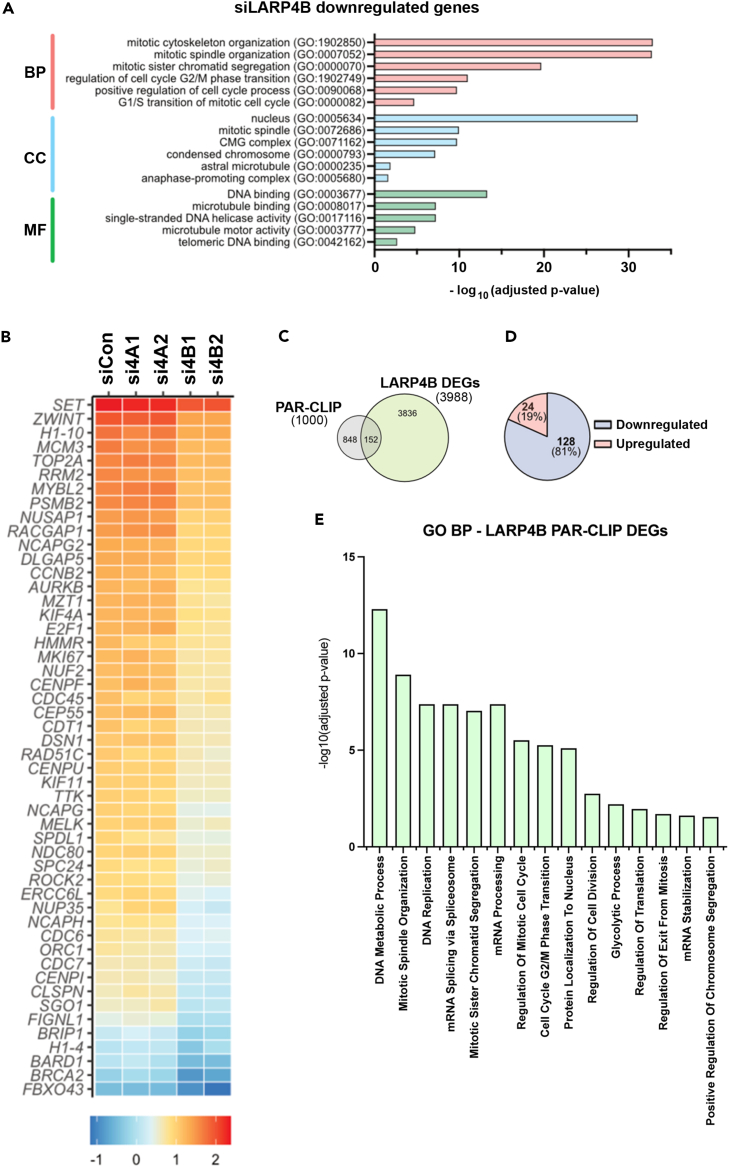
(A) Gene ontology (GO) terms for Biological Process (BP), Cell Component (CC), and Molecular Function (MF) significantly enriched for, following bulk RNAseq analysis of significantly downregulated genes in LARP4B knockdown PC3 cells compared to siControl cells.
(B) Heatmap depicting relative expression (bottom scale Z score) of the top 50 mitosis-related genes across siControl (siCon), siLARP4A (si4A1, si4A2), and siLARP4B (si4B1, si4B2) knockdown PC3 cells.
(C) Venn diagram showing overlap between mRNAs that feature as differentially expressed genes (DEGs) between LARP4B knockdown and control cells in bulk RNAseq, and candidate direct mRNA targets (top 1000) of LARP4B as shown by PAR-CLIP analysis by Kuspert et al.25
(D) Pie chart demonstrating the proportion of upregulated and downregulated DEG/PAR-CLIP genes that overlap with bulk RNAseq analysis of LARP4B knockdown PC3 cells compared to controls.
(E) Gene ontology (GO) terms for 15 of the top biological processes (BP) significantly enriched for, following analysis of significantly downregulated putative mRNA target genes of LARP4B, in LARP4B knockdown PC3 cells compared to controls. See also Figure S6.