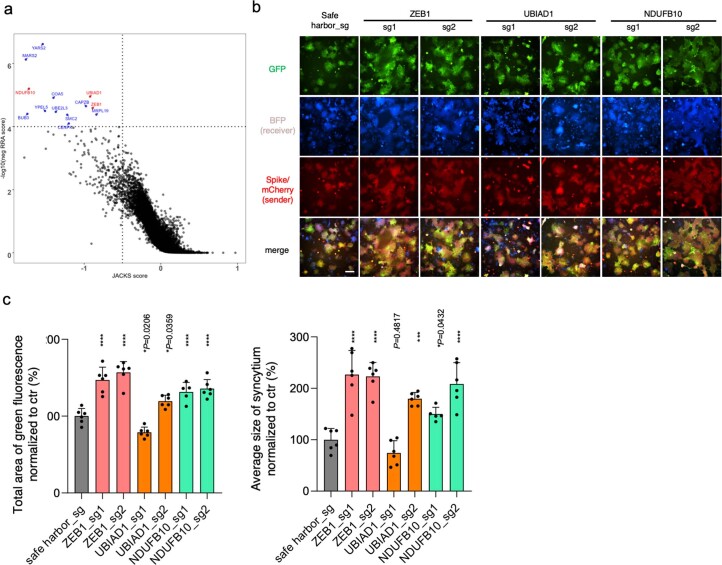
Extended Data Fig. 8. A genome-wide CRISPR screen reveals host-factor knockouts that enhance SARS-CoV-2 Spike-induced syncytium formation.
a) Depletion of sgRNAs in the unfused receiver cell population revealed by the RRA and JACKS scores. The sgRNA screen hits are highlighted in red, and the known essential genes are highlighted in blue. The genome-wide CRISPR screen data were collected from two biological replicates. b) Validation of screen hits using sgRNA-directed gene knockouts in A549-ACE2-Cas9-GFP1-10 receiver cells. The HEK293T-GFP-11 sender cells used express WT D614G Spike. Scale bar, 200 μm. c) Quantification of the syncytium area and average size of syncytium observed in (b). Data shown are mean ± SD (n = 6). P-values indicated were compared with safe harbor-targeting control. Statistical significance was determined using one-way ANOVA. n indicates the number of biological replicates. ***P < 0.001, ****P < 0.0001.