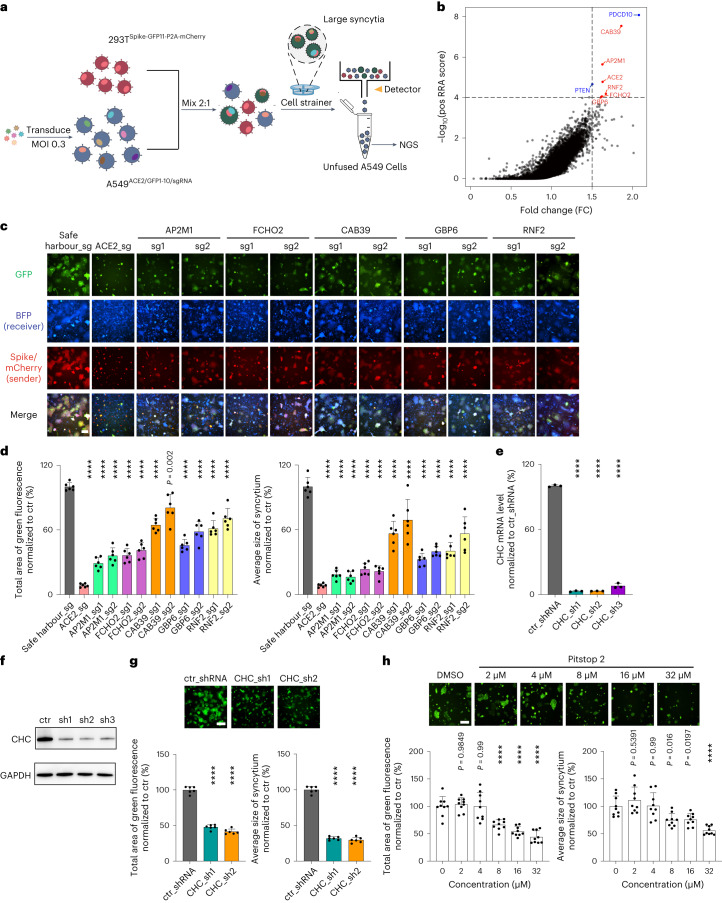
Fig. 4. Genome-wide CRISPR screen reveals host factors important for SARS-CoV-2 spike-induced syncytium formation.
a, Workflow for genome-wide CRISPR screening in receiver cells using a reverse selection approach. Large syncytia and small GFP+ syncytia were removed using cell strainer and FACS, respectively. Unfused A549-ACE2-SpCas9-GFP1–10-sgRNA cells were subjected to NovaSeq-based sequencing. b, Enrichment of sgRNAs in the unfused receiver cell population revealed by the RRA score and FC. FC represents each variant’s relative abundance in the unfused receiver cell pool vs the unmixed cell pool and is normalized to WT. The sgRNA screen hits are highlighted in red and the known tumour suppressor genes are highlighted in blue. The genome-wide CRISPR screen data were collected from two biological replicates. c, Validation of screen hits using sgRNA-directed gene knockouts in A549-ACE2-SpCas9-GFP1–10 receiver cells. The HEK293T-GFP-11 sender cells used express WT D614G spike. d, Quantification of the syncytium area (left) and average size of syncytium (right) observed in c. Data shown are mean ± s.d. (n = 6 biological replicates). e,f, Quantitative PCR with reverse transcription (RT–qPCR) (e) and western blotting (f) results on CHC knockdown. g,h, Inhibition of syncytium formation by CHC knockdown (g) and Pitstop2 treatment (h). GFP-split complementation system assay was applied as in c. Representative images are shown. Data shown are mean ± s.d. (n = 3 (e), n = 6 (g) and n = 9 (h) biological replicates). P values indicated were compared with safe harbour-targeting sgRNA (d), control (ctr) shRNA (e,g) or DMSO-treated control (h). Statistical significance was determined using one-way ANOVA. ****P < 0.0001. Scale bars, 200 μm.