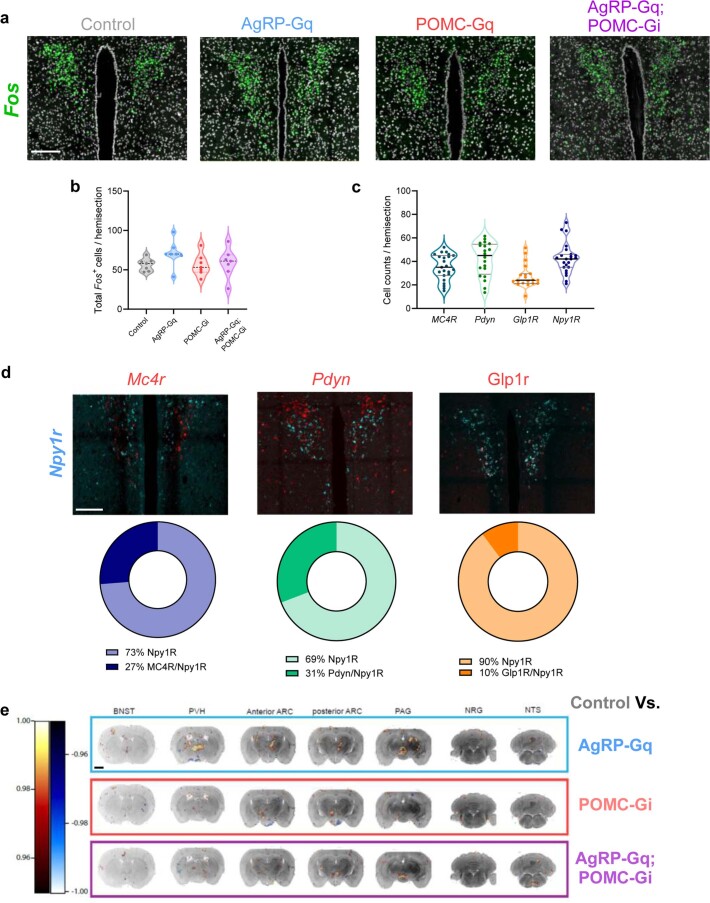
Extended Data Fig. 5. Distribution of PVH neuronal subpopulations and unbiased analysis of activated areas resulting from activation of AgRP- and inhibition of POMC neurons.
a, Representative images showing the distribution of Fos+ cells in the PVH after CNO treatment of the four experimental groups. b, Quantification of total Fos+ cells/hemisection in PVH area from CNO-treated mice for 1h. c, Quantification of total cells expressing Mc4r, Pdyn, Glp1r and Npy1r genes per PVH/hemisection area. d, Anatomical distribution and composition of Npy1R subpopulations that coexpress M, Pdyn, Glp1r markers in the medial PVH area, and quantification of percentage of total Npy1R+ population that coexpress another marker. e, Representative images of coronal sections across the whole brain highlighting areas with higher (yellow-white areas) or lower (blue-white) probability of containing Fos+ cells resulting from the statistical comparison of volumetric Fos distribution between control mice versus AgRP-Gq (top row, blue), POMC-Gi (middle row, red) and AgRP-Gq;POMC-Gi mice (lower row, magenta) after CNO treatment for 1h. Data represent average ± s.e.m of total cell counts per hemisection, from n = 7 mice/group in b,c, and from n = 4 mice/group in d, e. Statistical test: ordinary one-way ANOVA test for b. Source data and further details of statistical analyses are provided as a Source Data file. Scale bar = 100 µm.