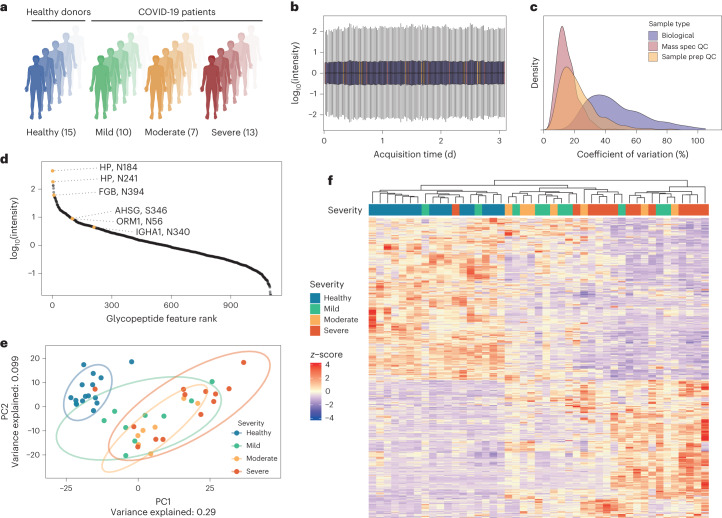
Fig. 3. Oxonium ion profiling allows robust and reproducible plasma glycoproteomics in a COVID-19 inpatient cohort.
a, COVID-19 inpatient cohort, comprising 30 patients hospitalized due to PCR-confirmed SARS-CoV-2 infection and 15 healthy controls. COVID-19 patients were distributed across different disease severities, ranging from mild (WHO 3), moderate (WHO 4, 5) to severe (WHO 6, 7) COVID-19. b, Total sample intensities across the MS measurement batch following median normalization; outliers are not plotted. Boxplot colours are the same as shown in panel c. c, Technical and biological variation across cohort measurements, indicated by distributions of c.v. values for glycopeptide features in repeat injections (mass spec QC, n = 10), commercial plasma (Tebu Bio) prepared in parallel with samples (sample prep QC, n = 9) and patient samples (n = 3 for each of 45 participants). d, Median intensity of glycopeptide features in a pooled sample, showing quantification spanning more than four orders of magnitude. Matched glycopeptide features are highlighted and labelled with their gene name and glycosite. e, PCA of all consistently detected features (n = 1,002) separates healthy and COVID-19 patients in PC1. The proportion of variation accounted for by each axis is shown in axis labels. f, Heat map and hierarchical clustering of differentially expressed glycopeptide features (calculated using the limma R package, |log2(fold-change)| > 1, adjusted P < 0.05) between COVID-19 patients and controls. Figure 3a created with BioRender.com.