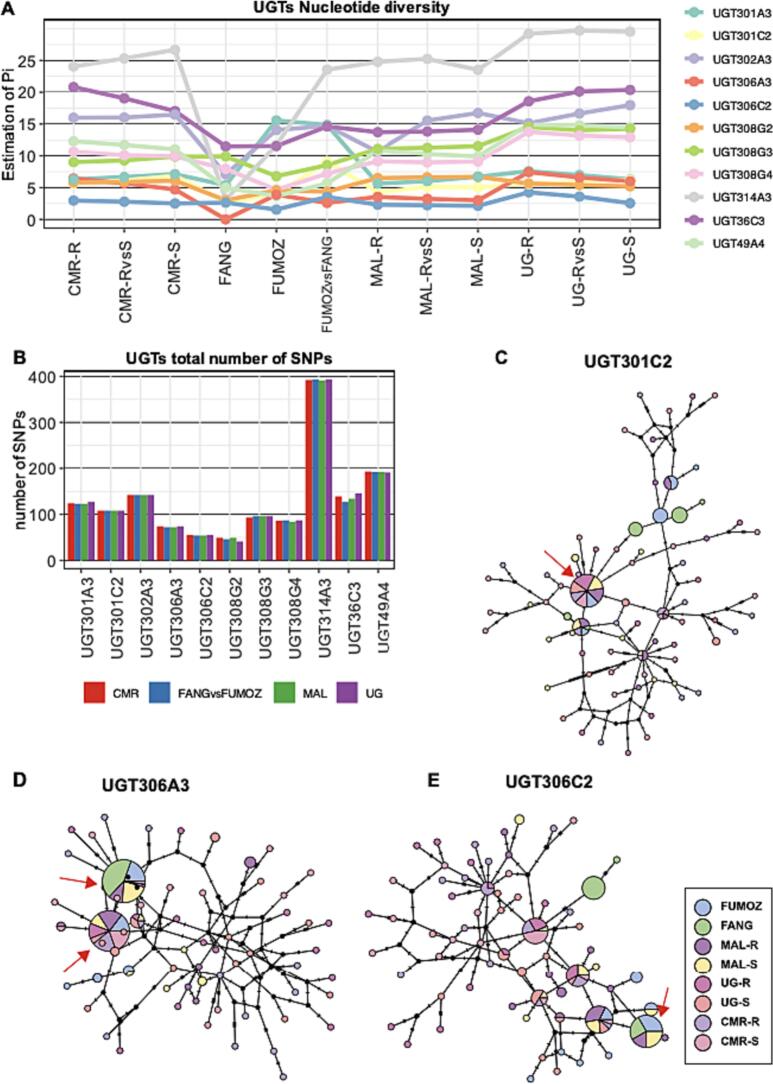
Fig. 5.
Targeted UGTs polymorphism and haplotype network analysis. UGTs gene-wise nucleotide diversity was calculated between (resistant and susceptible) populations and within populations from Malawi, Cameroon, Uganda and laboratory colonies the FANG and the FUMOZ (A). The number of polymorphic substitutions for UGTs in each country reflects the gene size (Table 1) (B). TCS haplotype network reveals limited haplotype clustering in UGT301C2 (C), UGT306A3 (D) and UGT306C2 (E) where predominant haplotypes shared by different populations were detected. The predominant haplotype in UGT301C2 was observed in (40/140) haplotypes from Cameroon (9), Uganda (14), Malawi (12) and FUMOZ (5). While 18/20 FANG haplotypes cluster together in three predominant nodes separated by a single mutation step each contains (9/20), (7/20), and (2/20) haplotypes respectively (Fig. 5C). In UGT306A3 two main haplotype clusters separated by a single mutation step were detected. The biggest cluster contains (45/160) haplotypes where all the 20 FANG haplotypes cluster, 9/20 FUMOZ and 15/40 haplotypes from Malawi. The second cluster contains (41/160) haplotypes including 12/40 from Malawi, 16/40 from Cameroon and 7/40 from Ugandan population (Fig. 5D). In UGT306C2 many haplotype clusters were observed the most dominant is 23 haplotypes cluster specific to FUMOZ (7/20), FANG (6/20) and Malawi (11/40).