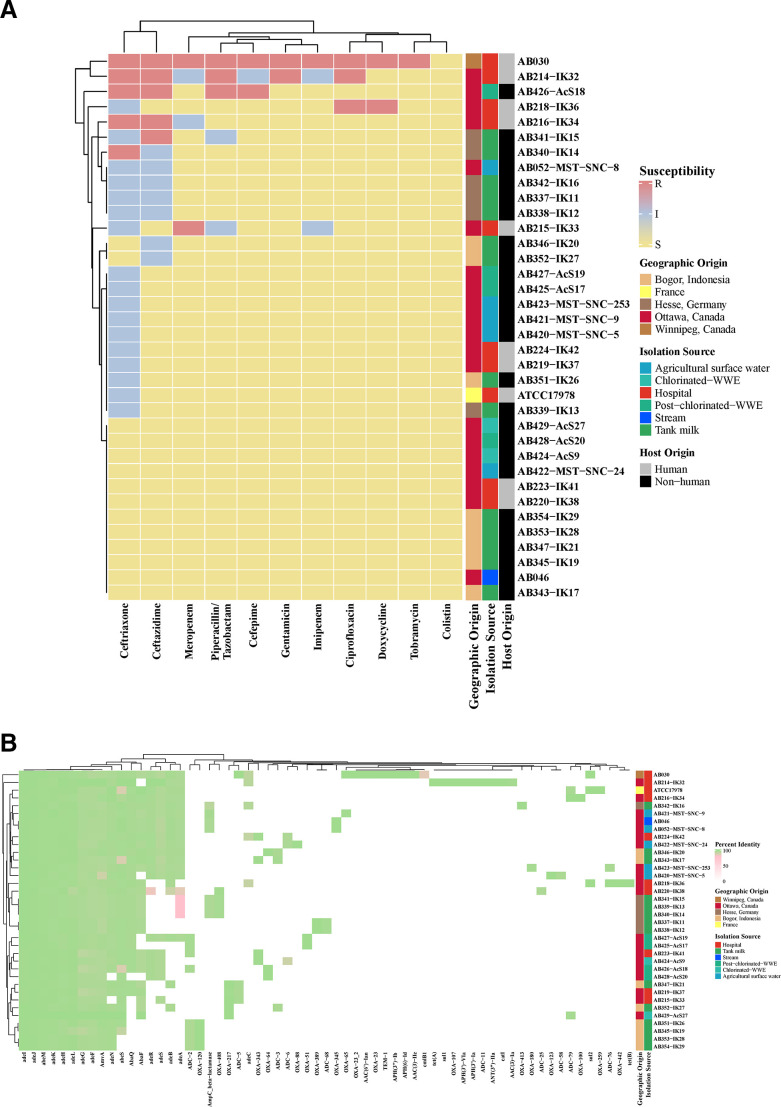
Fig 2.
(A) Antibiotic susceptibility profiles of all strains. Susceptibility testing was performed according to the CLSI guidelines. Clinically relevant antibiotics tested are part of the CANWARD panel and are as follows: amikacin, cefazolin, cefepime, cefoxitin, ceftazidime, ceftobiprole, ceftolozane/tazobactam, ceftriaxone, ciprofloxacin, clarithromycin, clindamycin, colistin, daptomycin, doxycycline, ertapenem, gentamicin, imipenem, linezolid, meropenem, nitrofurantoin, piperacillin/tazobactam, tobramycin, trimethoprim/sulfamethoxazole, and vancomycin. Only those antibiotics that showed at least one strain with a change in susceptibility based on CLSI breakpoints are shown. Color in each panel represents the isolation source and the geographic origin that corresponds to that of all figures. (B) Antibiotic resistance gene profile of all isolates. Percent identity of each gene is used to explore the diversity. The minimum cutoff for nucleotide identity is 80% and is shown in pink. Hierarchical clustering demonstrates similarity based on the antibiotic-resistant gene profile. Color in each panel represents the isolation source and the geographic origin that corresponds to that of all figures.