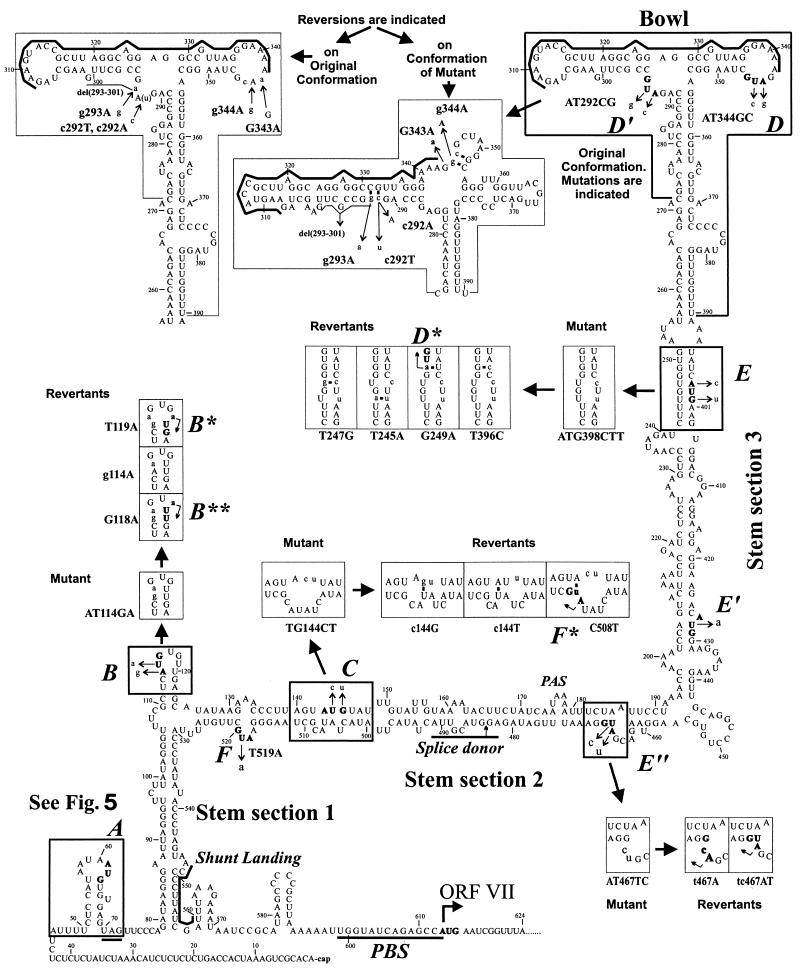
FIG. 4.
Reversions of the 35S RNA leader secondary structure and sORFs generated in planta. The MFold-predicted (20°C) hairpinlike secondary structure of the strain CM4-184 leader is shown. Here we refer to three parts of the main stem, which are divided by the side branches, as stem sections 1, 2, and 3 in ascending order and to the top of the structure as the bowl. Elements of this structure, which are influenced by the sORF mutations and the ensuing reversions, are boxed. The start codons of the sORFs are indicated in boldface and also by uppercase italic letters (A to F) along the sequence. Mutations are indicated by thin arrows. Mutant and revertant conformations of the structural elements are drawn in boxes nearby and indicated by thick arrows. Bent thin arrows within boxes show novel start codons created by the reversions in some cases (B*, B**, D*, F*). Nucleotides that do not occur in the wild-type sequence are in lowercase letters. Thick lines define the conserved purine-rich sequence of the bowl, the shunt landing sequence, the sORF A stop codon, the PBS, and the splice donor. PAS, polyadenylation signal.