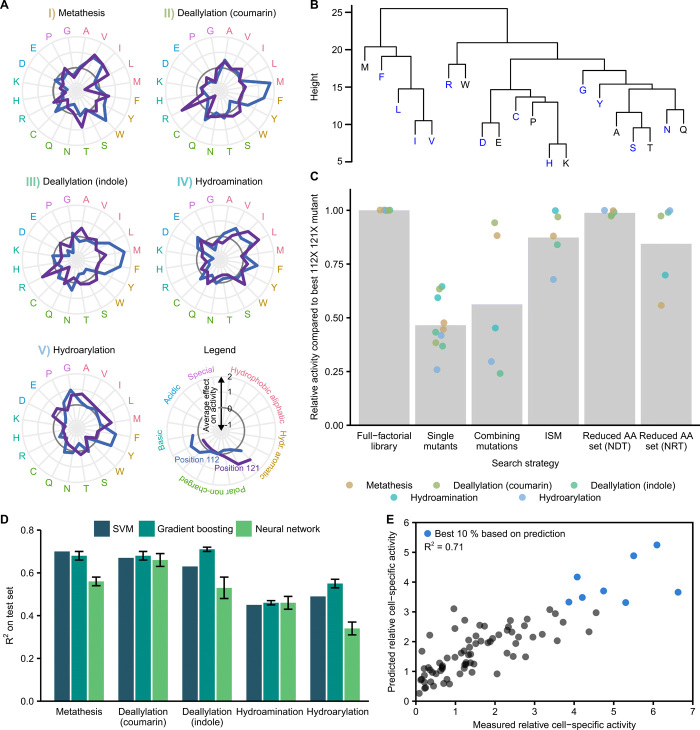
Fig. 4. Amino acid effects and implications for future ArM engineering.
(A) Effect of amino acids on ArM activity. Points outside and inside the dark gray circles indicate a positive and negative effect, respectively. Values were standardized by subtracting the mean activity of all 400 mutants and dividing by the corresponding SD. The mean across all 20 variants harboring the respective amino acid is shown. (B) Hierarchical clustering of amino acids across both positions and all reactions (Methods). NDT-encoded amino acids are highlighted (blue). (C) Comparison of enzyme engineering strategies. Strategies are compared to the full-factorial approach (left to right): screening single mutants (both positions), combining the best single mutations at both positions, ISM, and combinatorial screening with reduced amino acid sets (NDT/NRT codons). Bars are the mean across hits identified by the respective strategy. (D) Predictive machine learning models based on SVM, gradient boosting, and neural networks (Methods). Models were trained on 320 randomly selected data points and evaluated on the remaining 80 variants. Mean and SD of five training runs are displayed for gradient boosting and neural networks. (E) Performance of gradient boosting model for reaction III. Predictions are plotted against experimental measurements for 80 held-out variants (predicted top 10% highlighted in blue).