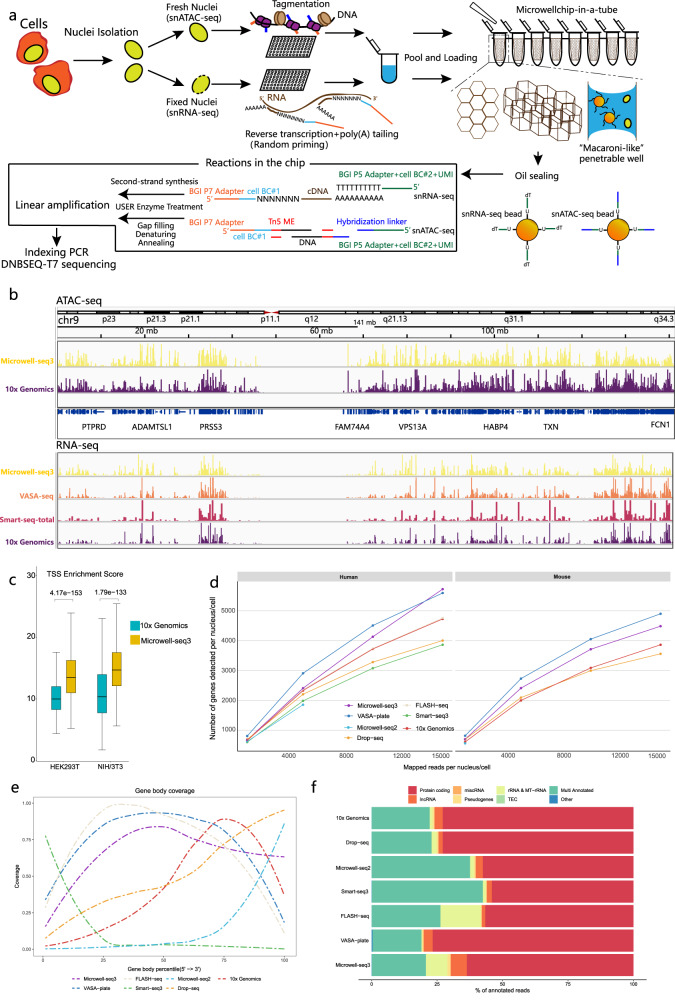
Fig. 1. Overview of the microwell-seq3 workflow.
a Schematic view of the Microwell-seq3 workflow. Single nuclei are extracted from fresh or frozen tissues. DNA or RNA in nuclei is tagged with the first part of the cell barcode (BC#1) in multiple 96-well plates. Labeled nuclei and barcoded magnetic beads are pooled and evenly loaded into Microwell chips. Oligos are released from the beads and linear preamplification is performed in the chips. Preamplified DNA or RNA fragments are collected from the chips for final library preparation and sequencing. b Representative University of California Santa Cruz (UCSC) Genome Browser view of ATAC-seq and RNA-seq signal tracks (HEK293T cells) from Microwell-seq3, 10X Genomics, VASA-seq and Smart-seq-total data. c Distribution of TSS enrichment scores from Microwell-seq3 and 10X Genomics ATAC-seq data in the indicated cell lines, reads (fragments) are down sampled to 2000 fragments per nucleus. The statistical test used is a two-sided Student’s t-test. d Number of the detected annotated genes in human and mouse cell lines (HEK293T, mouse NIH/3T3 and embryonic stem cells (mESCs)) in each method is plotted against the number of unique mapped reads per cell in different down-sampling thresholds. All the reads are remapped with the same pipeline after adapter filtering and trimming. Data of mESCs were generated by VASA-seq. e Comparison of mean ± standard deviation (SD) gene body coverage in protein-coding genes in cell lines across the different methods. Microwell-seq3, FLASH-seq and VASA-seq show even coverage across the length of the genes. Other methods show bias toward the 5′ and 3′ ends of transcripts, respectively. Reads in each method are trimmed to 500,000 per sample. f Proportions of the reads mapped to all annotated genes for each biotype in cell lines across the different methods. Microwell-seq3 detects proportionally higher levels of lncRNAs. Reads in each method are trimmed to 5000 per cell.