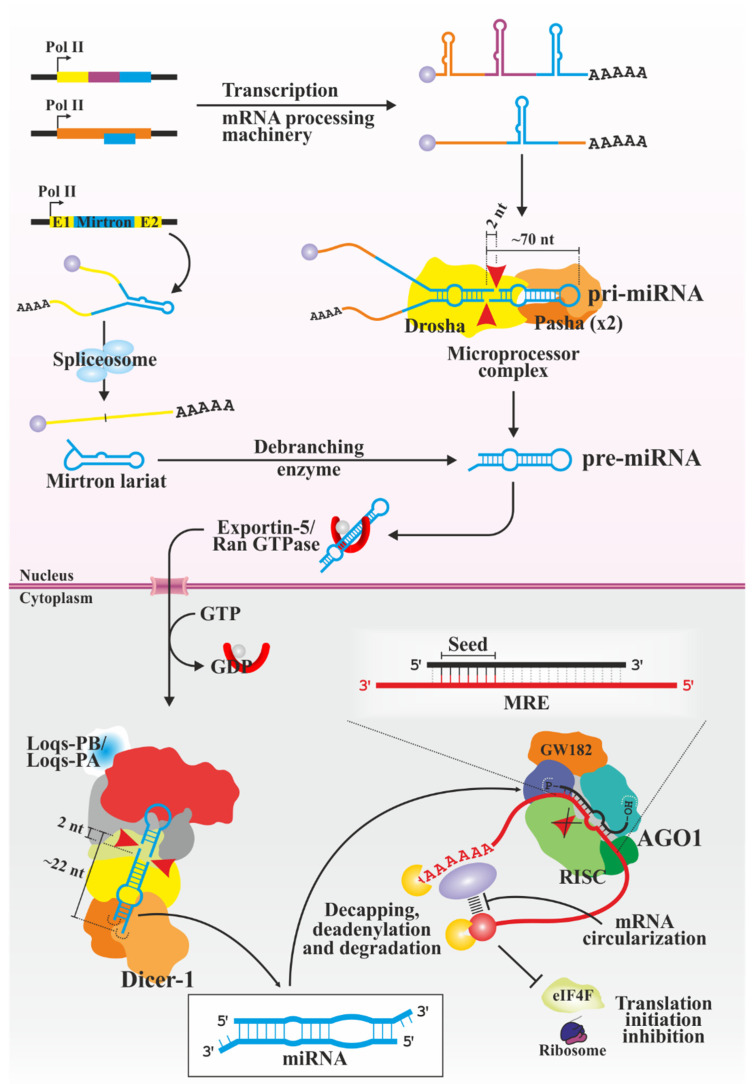
Figure 2.
miRNA pathway in insects. miRNA-containing genomic loci are transcribed by RNA polymerase II generating long, partially dsRNA primary precursors (depicted in blue) that are trimmed into shorter hairpins by the Microprocessor complex Drosha/Pasha in the nucleus. This step is not necessary in the case of mirtrons, whose processing depends on spliceosomal and debranching machinery. The precursors are then exported into the cytoplasm where Dicer-1, aided by Loqs isoforms, eliminates the loop resulting in ~22 bp miRNA. The characteristic cleavage of the Microprocessor and Dicer-1 are shown. The miRNA guide strand (black) loaded into AGO1 induces cleavage-independent mRNA (in red, depicted with 5’ cap and 3’ poly(A) tail) degradation and translation suppression of the partially complementary mRNAs. -OH and -P, 3′-hydroxyl and 5-phosphate termini, respectively; AGO1, Argonaute 1; E1/E2, exon 1 and 2, respectively; eIF4F, eukaryotic initiation factor 4F; Loqs-PA/-PB, Loquacious isoforms PA and PB, respectively; miRNA, micro RNA; MRE, microRNA recognition elements; Pol II, RNA polymerase II; pre- and pri-miRNA, precursor and primary micro RNA, respectively; and RISC, RNA-induced silencing complex.