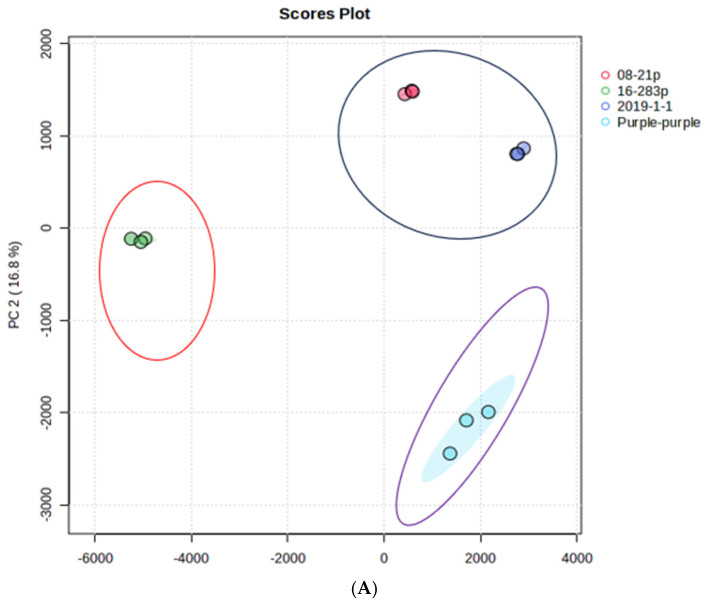
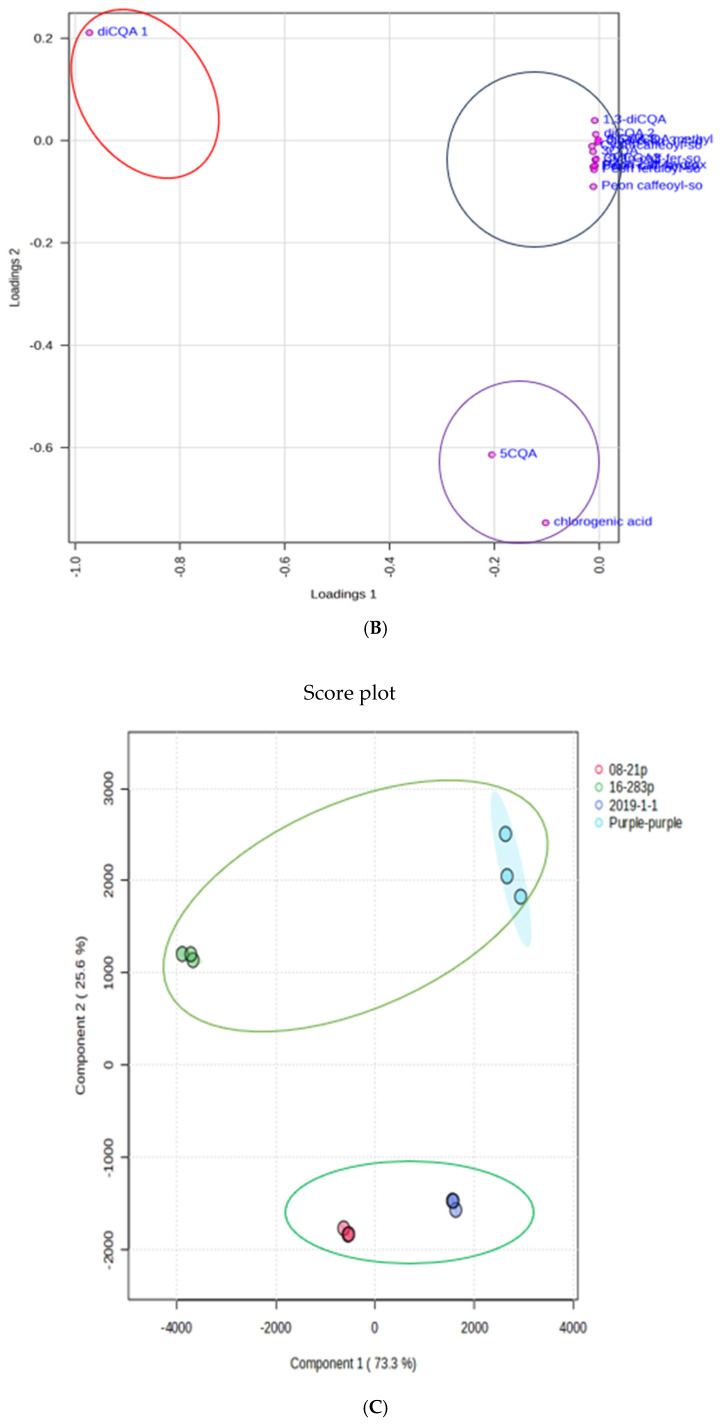
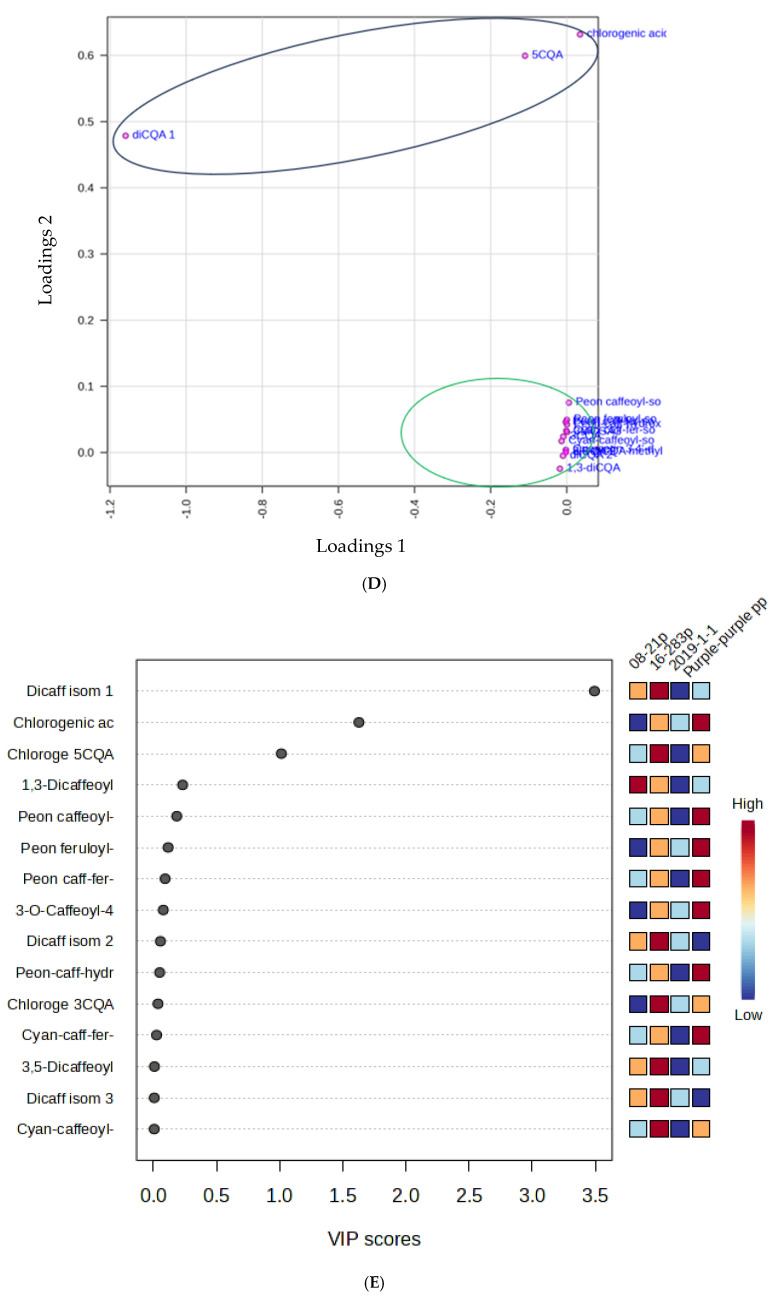
Figure 2.
(A) An unsupervised PCA score plot of phenolic metabolites generated by UPLC-QTOF/MS analysis showing the separation of three clusters. (B) Loading of phenolic metabolites in the PCA score plot. (C) A PLS-DA score plot showing four sweet potato cultivars clustered into three groups. (D) PLS-DA score plots loaded with different phenolic compounds detected by UPLC-QTOF-MS showing two clustered groups, (E). Metabolites are assigned VIP scores in PLS-DA. Variable importance is determined by the score they receive from low to high. Each metabolite’s relative concentration is shown in the coloured boxes on the right. Low blue levels indicate low levels, while high red levels indicate high levels. (F) Heat map. In the map, the various phenolic compounds found in different sweet potato cultivars are coloured according to their concentration. The rows represent phenolic compounds, and the columns represent the sweet potato genotype. The colours red and blue indicate high and low levels, respectively.