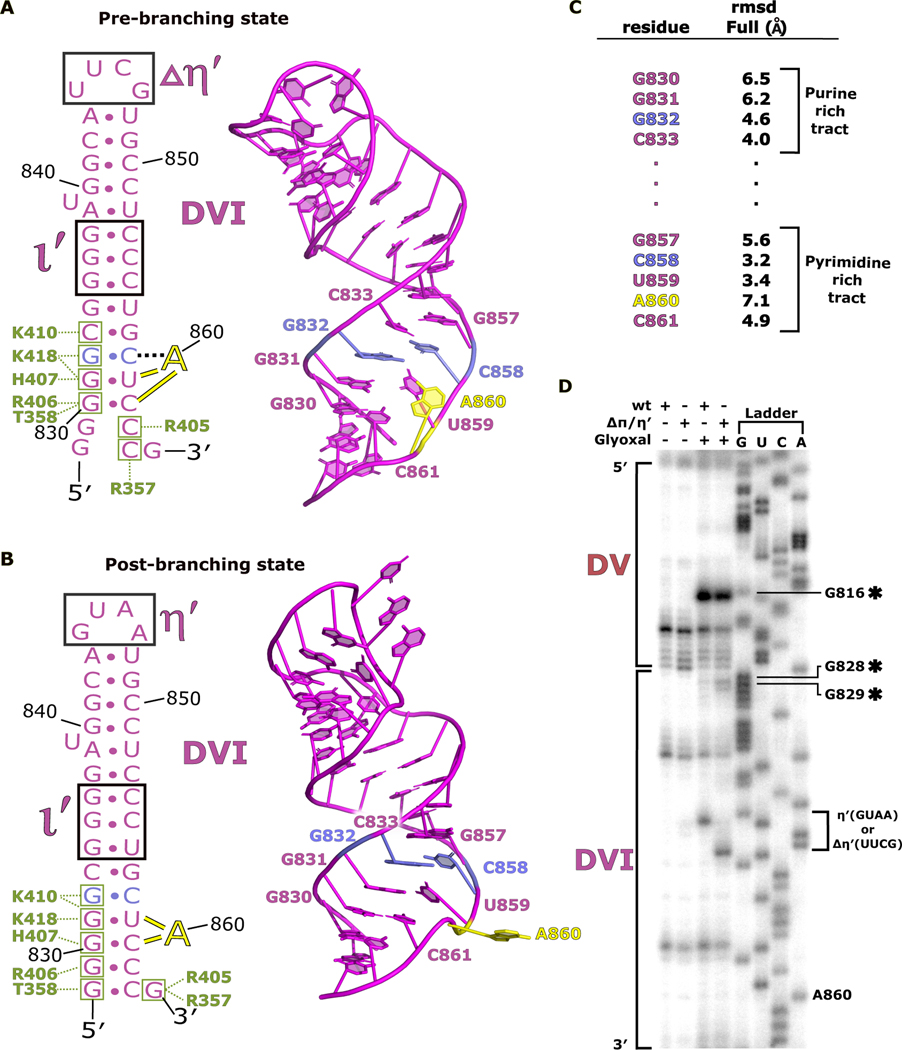
Fig 5. DVI helix undergoes remodeling during branching.
a. The secondary structure and atomic model for DVI (magenta) is shown for the pre-branching state. The branch site adenosine A860 (yellow) is forming the base triple with G832 and C858 (light blue) prior to branching. The matX-DVI (green boxes and corresponding amino acid labels) and ι-ι′ (black box) are holding DVI in the horizontal conformation required for branching. b. The secondary structure and 3D model for DVI is shown for the post-branching state. A860 has rotated out of the DVI helix removing the lariat bond from the active site. The new footprint for the matX-DVI and ι-ι′ interactions is shown highlighting the dynamic movement of DVI during branching. c. Full residue RMSD values are reported for the nucleotides surrounding the branch site. d. Glyoxal probing was performed to evaluate the stability of the DVI helix during branching. The appearance of glyoxal modification at G828 and G829 in the ΔπΔη′ mutant suggest that the base of the DVI helix is dynamic during branching.