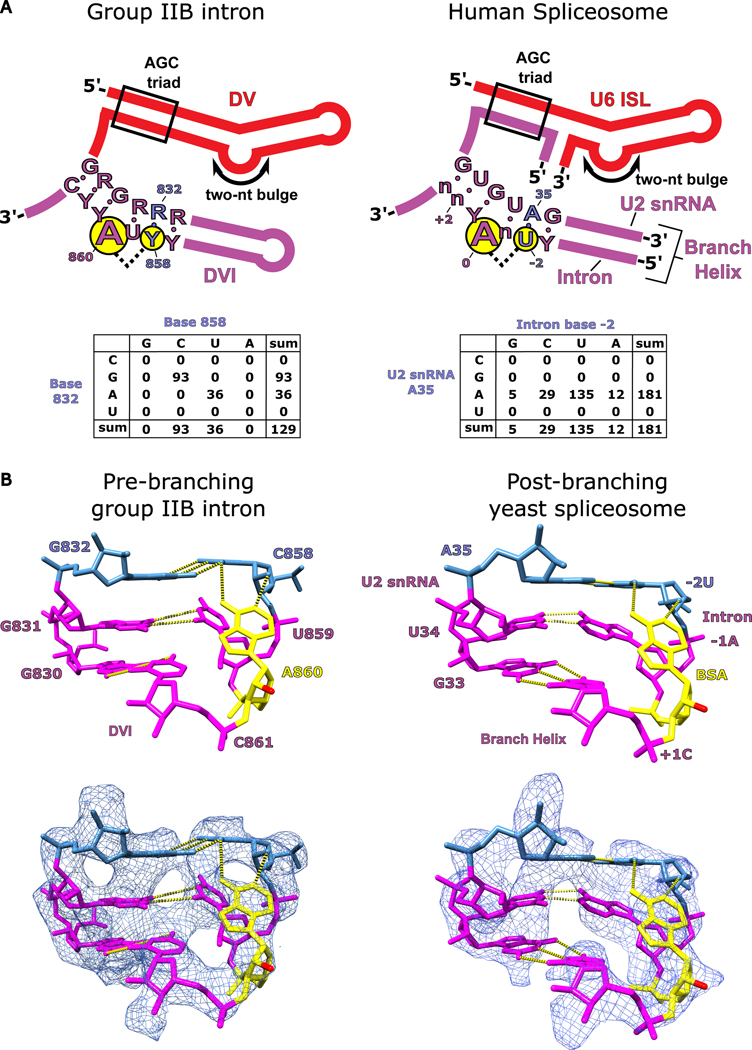
Figure 7. Conservation of branch-site helix architecture.
a. The consensus sequence for DV and DVI of group IIB introns is shown alongside the homologous U6 ISL/U2 snRNA complex and branch helix of the human spliceosome. The structural requirements to form the base triple (G832-C858-A860) involved in branching for the group II intron are highly conserved and the conservation is maintained within the branch helix of the human spliceosome. A covariation analysis of the base triple in DVI of the group II intron shows a complete Watson-Crick requirement for nucleotides 832 and 858. Covariation of the branch helix within the human spliceosome shows a strong A-U base pair preference for the nucleotides forming the base triple with the branch-site adenosine. b. The hydrogen bonding network responsible for positioning the branch-site adenosine (A860) in the pre-branching state is shown for the T.el4h group IIB intron. The 2′-OH nucleophile is shown in red and has been extruded from the branch helix poised to attack the 5′ SS. A previous structure shows the branch helix for a post-branching spliceosome from yeast captured in the C complex immediately after lariat bond formation (PDB: 5LJ3)4. The analogous region of the branch helix in the spliceosome is shown and has an almost identical structural geometry to that observed in the group II intron (human U2 snRNA numbering was used for figure clarity). The base triple in the spliceosome occurs between the branch-site adenosine (BSA), A35 of the U2 snRNA, and the −2U of the intron. This structural conservation reveals that the fundamental basis for how the branch-site adenosine is positioned has remained unchanged over billions of years of evolution.