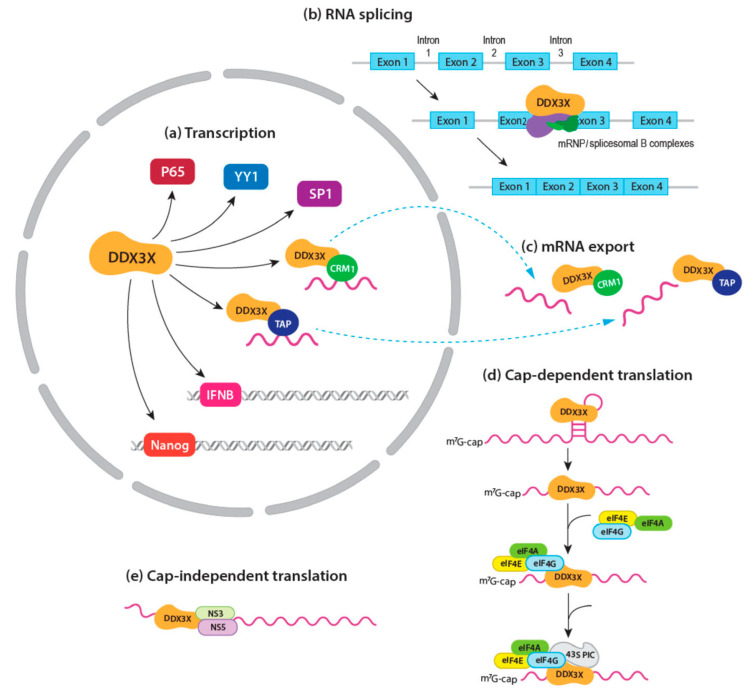
Figure 2.
Roles of DDX3X in mRNA metabolism. (a) DDX3X regulates transcription through its interactions with transcription factors such as p65, Yin Yang 1 (YY1), and SP1. DDX3X can also directly bind to IFNB and Nanog promoters to manipulate transcription. (b) DDX3X has been identified in human messenger ribonucleoproteins (mRNPs) and spliceosomal B complexes in physiological environments, which are involved in mRNA splicing. The green and purple shapes indicate DDX3-binding molecules in mRNPs and spliceosomal B. (c) DDX3X promotes mRNA export via binding to mRNA-exporting proteins CRM1 and Tip-associated protein (TAP). (d) DDX3X is able to unwind the secondary structure of mRNA, and promotes ribosome access to mRNA during cap-dependent translation. Currently, it is widely accepted that the translational regulation of DDX3X is mediated by eukaryotic translation initiation factors (eIFs). (e) Cap-independent translation usually occurs under stress conditions or during viral infection. In Japanese encephalitis virus (JEV) infection, DDX3X is bound to JEV non-structural protein 3 (NS3), NS5, as well as viral RNA.