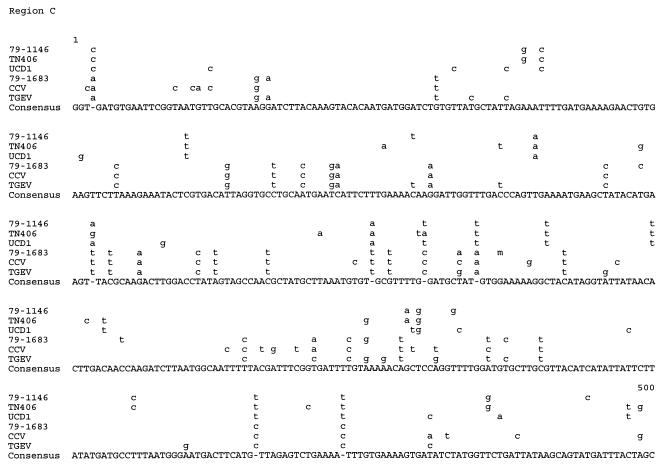
FIG. 3.
Comparative sequence analysis of genomic regions A through D. Alignments were made by using the PILEUP program (University of Wisconsin), which scores identity between every possible pair (17). Alignment of the nucleotide sequences of regions A through D, as determined for the FCoV type I strains UCD1 and TN406, the FCoV type II strains 79-1683 and 79-1146, the CCV strain K378, and the TGEV strain Purdue (11), is shown. Only those nucleotides differing from the consensus sequence are depicted. n, r, and w represent nucleotides A/C/G/T, A/G, and A/T, respectively.