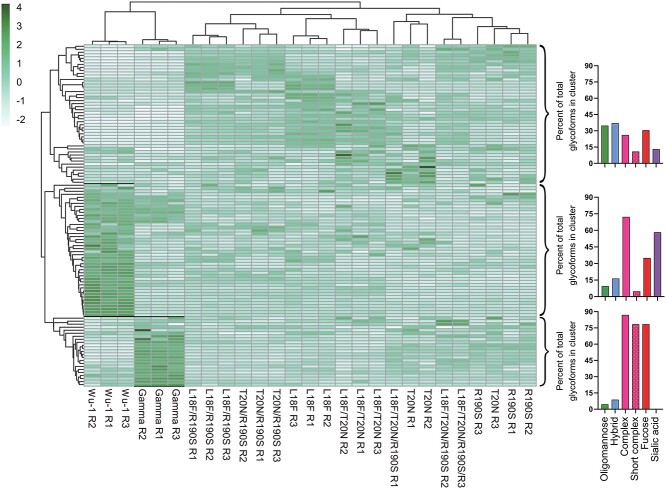
Fig. 5.
SARS-CoV-2 spike proteins of Wu-1, gamma and NTD mutants have diverse glycosylation profiles. Clustered heatmap of the relative abundance of all identified glycoforms in all replicates, excluding N188. Both rows and columns were clustered using correlation distance and average linkage. The percentage of types of glycans observed in each selected cluster are represented in bar graphs (refer to the methods for the allocation of glycan-types). The full clustered heatmap with labelled rows (glycopeptide glycoforms) is shown in Supplementary Fig. 3A.