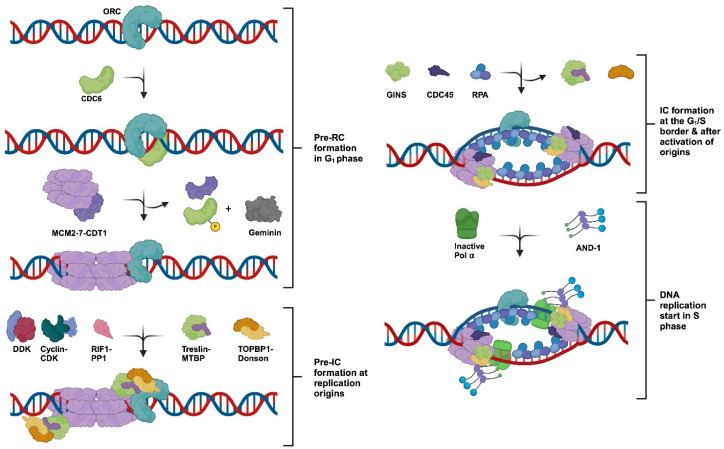
Figure 1.
The initiation process at eukaryotic origins of DNA replication. At the end of mitosis and in the early G1 phase of the cell cycle, ORC, the origin-recognition complex, binds to eukaryotic origins of replication together with CDC6. In G1, CDT1 (chromatin licensing and DNA replication factor 1) together with the MCM2-7 complex then associates with the CDC6–ORC complex, and the MCM2-7 proteins are loaded as helicase-inactive double hexamers (MCM-DHs) onto the chromatin, forming the pre-replicative complex (Pre-RC) and license the origin. The modification of CDC6 and binding of CDT1 to Geminin inactivates the loading activity of these proteins with CDC6 being degraded similarly as free CDT1. In the next step, Treslin-MTBP (SLD3-SLD7 in yeast) interacts with MCM-DHs at the chromatin, and the DBF4/DRF1 CDC7 kinase (DDK) phosphorylates the MCM2-7 proteins. The DDK-dependent phosphorylation can be reversed by RIF1-PP1 making this step reversible. Next, Cyclin-CDKs phosphorylate Treslin and stimulate the formation of Donson–TOPBP1 complexes, which in turn bind to MCM-DHs. Donson–TOPBP1 supports the loading of the GINS complex and its association with MCM-DHs. The binding of CDC45 leads to the formation of the CMG complex and its activation, whereas TOPBP1 and Treslin–MTBP are released from the chromatin. CryoEM data suggest that Donson associates as a dimer with CMG, but only one Donson subunit binds to GINS and MCM2-7 proteins stabilising the CMG complex [28]. In the following step, two replication forks (RFs) are formed and replication protein A (RPA), with the help of the CDC45, binds to and stabilises the resulting ssDNA. The association of AND-1/CTF4/WDHD1 (shown as AND-1 in the diagram) with CMG allows for the loading of an inactive DNA polymerase α (Pol α) (dark green), including its primase subunits, to RFs. The activation of Pol α (light green) permits the primase subunit PRIM1/PRI1, with the help of PRIM2/PRI2 and additional replication factors, to synthesise the first RNA primer in origin sequences, resulting in the completion of the initiation process at origins and the start of the elongation phase. Additional proteins associated with RFs, such as the fork-stabilising proteins Timeless, Tipin, and Claspin plus Pol ε [3,42,43], were omitted in the diagram for simplification and clarity reasons providing a better overview. Adapted using information from [3,26,27,28,44,45,46] and created with BioRender.com.