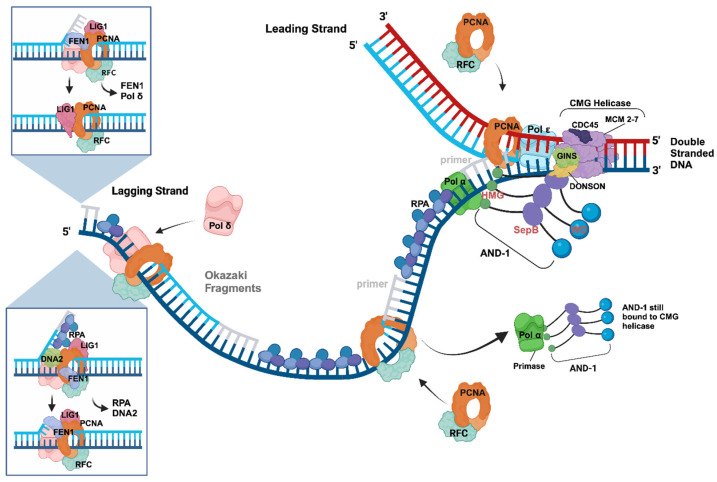
Figure 2.
Leading and lagging strand synthesis at a eukaryotic replication fork. In this RF model, CMG helicase (CDC45-MCM2-7-GINS with CDC45, in dark blue, the MCM2-7 hexamer, in purple, and GINS, in light green) unwinds the parental dsDNA into the leading and lagging strand templates (dark-red and dark blue, respectively). The protein Donson associates with the CMG complex during its formation and remains attached to it during the unwinding reaction. Additionally, the diagram shows the replication proteins that are involved in DNA synthesis and the maturation of Okazaki fragments. As seen in the model, RPA heterotrimers (three shades of blue) bind to the unwound ssDNAs preventing hairpin formation and nuclease-dependent ssDNA degradation. RFC (blue) loads the PCNA ring (red brown) onto the primed template DNA. The latter stabilises Pol ε (light blue) on the template DNA when synthesising the leading strand (light blue DNA). Pol ε also associates with the CMG complex to support its unwinding activity, but this interaction might also be important during replication fork stalling (see Section 5). For lagging strand DNA synthesis, the AND-1/CTF4/WDHD1 homotrimer (named AND-1 in the diagram with one subunit consisting of an HMG (green), SepB (dark blue), and WD (blue) domain) links CMG to the Pol α complex (green). The primase function of Pol α synthesises the RNA primer (light grey), starting Okazaki fragment synthesis during lagging strand synthesis. After the initiation step, primase hands over the RNA primer to the DNA polymerase domain of Pol α on PolA1 (first polymerase transition). The latter extends the RNA primer and synthesises a short RNA–DNA fragment before leaving the template. RFC (blue) replaces Pol α with the help of RPA and loads PCNA, the DNA clamp, on the primed DNA. This RFC–PCNA complex allows Pol δ (pink) to associate with the RNA–DNA primer (2nd polymerase transition). The RFC–PCNA–Pol δ complex elongates this RNA–DNA in a processive manner until it reaches the next Okazaki fragment. Then, Pol δ slows down but continues to elongate the newly synthesised DNA. The polymerase displaces the RNA and parts of the Pol α-synthesised DNA of the Okazaki fragment in front (strand displacement). Thus, Pol δ produces an RNA-DNA flap, which is recognised and cleaved by PCNA-associated FEN1 (blue), creating a perfect product, nicked DNA, for LIG1 (top panel on the left; the two inserted panels provide insights into the different pathways of the Okazaki fragment maturation process). The DNA ligase LIG1, which is also bound to PCNA along with Pol δ and FEN1, then ligates the two DNA fragments, yielding a continuous stretch of DNA. In an alternate pathway, RPA binds the flap structure produced by Pol δ competing with FEN1 (lower inserted panel). RPA recruits the DNA2 helicase/endonuclease to the flap structure. The latter in turn cleaves the ssDNA but leaving an extra nucleotide remaining, which results in a product that LIG1 does not ligate. After RPA and DNA2 have left the DNA, FEN1 cuts off the remaining base and LIG1 ligates the two DNA fragments. Adapted from [2,32,61,67] and created with BioRender.com.