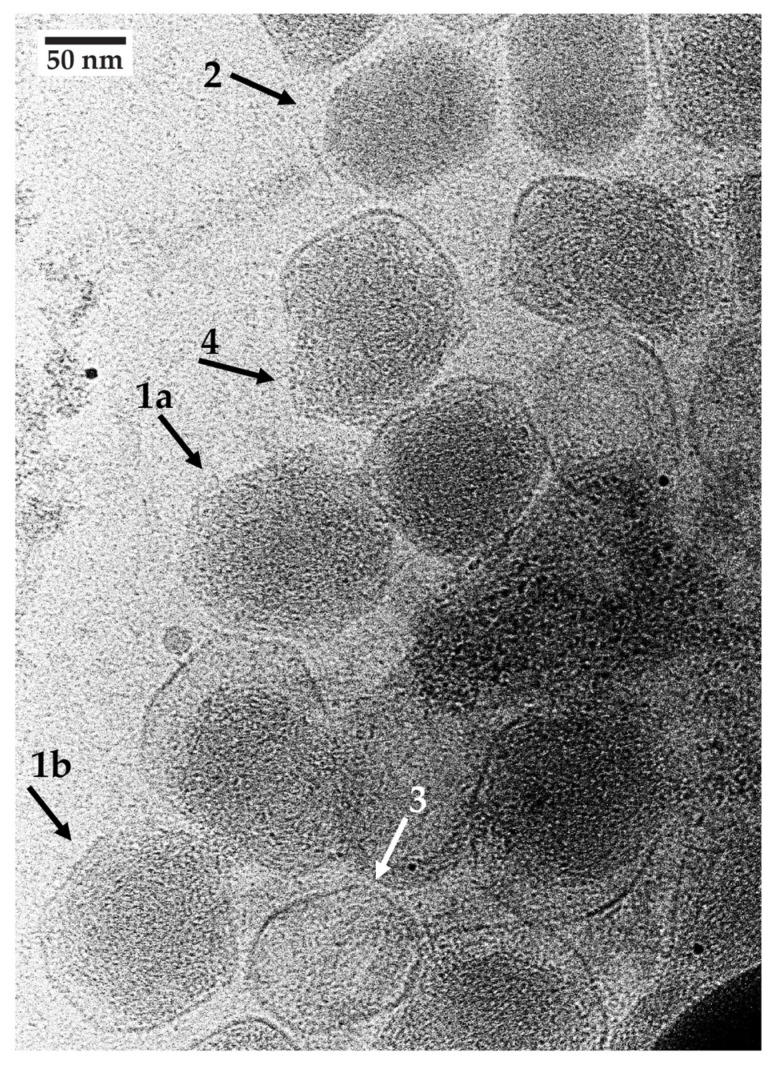
Figure 4.
Cryo-electron microscopy of MB-reacted, unstained phage T4. Phage T4 was incubated with 10.0 mM MB as in Figure 2 and then immediately prepared for cryo-EM by use of the following procedure. The support was a Quantifoil copper holey carbon grid (R3.5/1, 200 mesh, Electron Microscopy Sciences, Hatfield, PA, USA) with a 2 nm ultrathin carbon layer. Grids were glow-discharged at 20 mA for 30 s in a Quorum EMS glow discharge machine (Iowa City, IA, USA). Then, a 3 ul phage sample was applied to a grid. The samples were blotted for 3 s with a blot force of 10 with filter paper under 100% humidity and a temperature of 8 °C in the Vitrobot Mark IV (Thermo Fisher Scientific, Waltham, MA, USA) chamber. Grids were plunge-frozen in liquid ethane prior to immediate transfer into liquid nitrogen for storage. Imaging was performed with a 200 kV Glacios cryo-TEM microscope (ThermoFisher Scientific, Waltham, MA, USA) with a gun lens of 4 and a spot size of 7. A 50 uM C2 aperture was used to adjust a parallel beam. All of the beam parameters were adjusted with a Falcon4 camera plus Selectris energy filter (ThermoFisher Scientific) by using Thermo Fisher Scientific EPU software, version 3.7. The beam parameters were the following: Atlas 110 X, Square 540 X, Hole 7600 X. The image acquisition magnification was 63,000 X with a pixel size of 1.89 Å, field of view of 775.4 nm and 10 eV slit width. After the grid Atlas map acquisition, a target-thinner ice square was chosen to map the whole picture, where target holes were used for the final data. Image acquisition was performed with MRC image file format. Images were recorded with an exposure time of 10 s, total dose of 20.22 e−/Å2 and dose rate of 2.02 e−/pix/s and the defocus range was manually adjusted from −2.5 to −2.0.