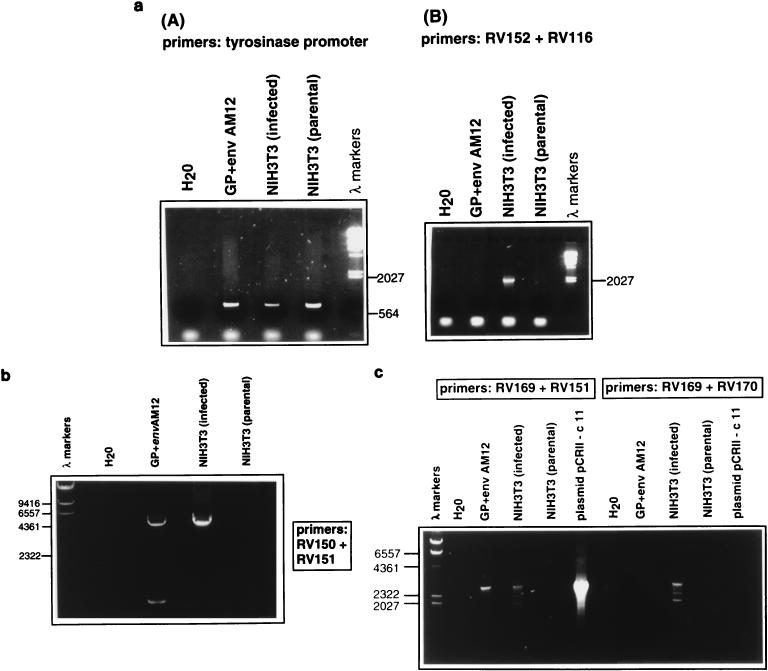
FIG. 3.
(a) Amplification of an env-3′ LTR fragment by PCR with DNA from helper virus-infected NIH 3T3 cells. (A) As controls for the presence of genomic DNA, PCR was performed with primers for the promoter of the murine tyrosinase gene, with DNA of GP+envAM12 cells and parental and helper virus-infected NIH 3T3 cells. (B) An env-LTR fragment was amplified with primers RV152 plus RV116. A positive signal was obtained only in the reaction with DNA of infected NIH 3T3 cells. (b) Amplification of a 5′ LTR–gag-pol fragment by PCR with DNA from helper virus-infected cells. Primer RV150 corresponds to a sequence in MoMLV LTR, and RV151 is at the pol/env overlap region. PCR with DNA of GP+envAM12 showed two signals, consistent with amplification from the two packaging constructs. The reaction with DNA of infected NIH 3T3 cells showed one signal of approximately 5.7 kb, which was absent in the reaction with parental NIH 3T3 cells. (c) Amplification of a pol-4070A env fragment by PCR with DNA from helper virus-infected cells. For PCR with primers RV169 plus RV170, several signals, the largest and most prominent of which was approximately 2.7 kb, were obtained in the reaction with DNA of infected NIH 3T3 cells. These were absent in the other samples. For PCR with primers RV169 plus RV151, signals were present in the reactions with DNA from infected NIH 3T3 cells and GP+envAM12. A positive signal was also obtained with plasmid pCRII-c11, which bears the cloned PCR-amplified fragment obtained from the reaction shown in panel b.