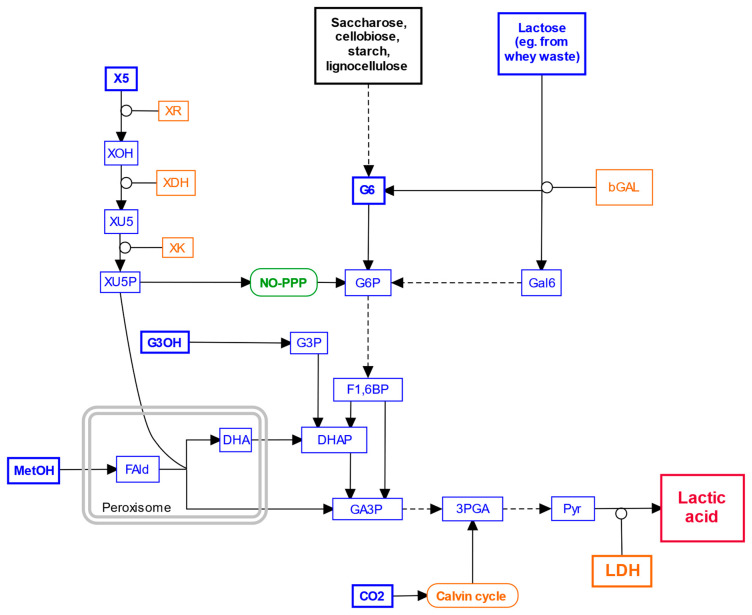
Figure 2.
Simplified scheme of pathways leading to lactic acid from various carbon sources reported to have been used in yeast and filamentous fungi. Blue are individual metabolites, bolded for the ones fed to the target organisms in cultures; the exception is lactic acid, which is red for emphasis. Black are miscellaneous carbon sources that feed into the same pathways. Orange are enzymes and pathways that may need to be introduced as transgenes into the target organisms for them to be able to use a given carbon source; LDH is bolded for emphasis. Green is a native pathway not shown in detail for simplicity. Co-substrates, coproducts, transporters, and competing pathways are omitted for clarity. Solid arrows are single, and dashed arrows multiple enzymatic steps. Metabolite key: 3PGA, 3-phosphoglycerate; CO2, carbon dioxide; DHA, dihydroxyacetone; DHAP, dihydroxyacetone phosphate; F1,6BP, fructose 1,6-bisphosphate; FAld, formaldehyde; G3OH, glycerol; G3P, glycerol 3-phosphate; G6, glucose; G6P, glucose 6-phosphate; GA3P, glyceraldehyde 3-phosphate; Gal6, galactose; MetOH, methanol; Pyr, pyruvate; X5, xylose; XOH, xylitol; XU5, xylulose; XU5P, xylulose 5-phosphate. Enzyme key: bGAL, beta-galactosidase (EC 3.2.1.23); LDH, lactate dehydrogenase; XDH, xylitol dehydrogenase (EC 1.1.1.B64); XK, xylulokinase (EC 2.7.1.17); XR, D-xylose reductase (EC 1.1.1.307). NO-PPP, non-oxidative phase of the pentose phosphate pathway. See text for details. Made with PathVisio.