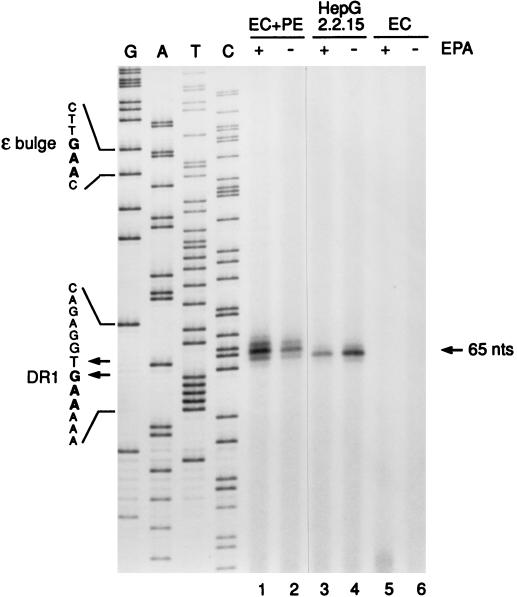
FIG. 4.
Primer extension analysis of Pol-associated DNA isolated from recombinant HBV nucleocapsids reveals DR1 as the origin of minus-strand DNA elongation. Following in vitro polymerase reactions in the presence (+ EPA) or absence (− EPA) of deoxynucleotides, covalently linked minus-strand DNA-Pol adducts were recovered from immunoprecipitated HBV capsids from EC+PE (lanes 1 and 2)- or EC (lanes 5 and 6)-expressing Sf9 cell lysates as described in Materials and Methods. Intracellular cores from 2.2.15 cells (lanes 3 and 4) served as a positive control. Purified DNA was annealed with a 5′-end-labeled plus-polarity oligonucleotide spanning nt 1764 to 1783 and then extended with a thermostable (exo-minus) polymerase and PCR cycling. Extension products were resolved on an 8% polyacrylamide sequencing gel along with a plus-strand dideoxy sequencing ladder (lanes G, A, T, and C) generated with the same primer and recombinant HBV DNA. The 65-nt extension product mapping to DR1 is indicated on the right. Shown on the left are minus-strand DNA sequences derived from the indicated portions of the plus-strand sequence ladder; these sequences include parts of the priming site in the bulge of 3′ ɛ and the elongation site in DR1. Minus-strand DNA 5′ ends mapping to nt 1829 (T residue) and 1828 (G residue) within DR1 are also indicated (arrows). Note that the primer extension products migrated approximately 0.5 to 1.5 nt more slowly than the corresponding position indicated by the sequencing ladder (see text).