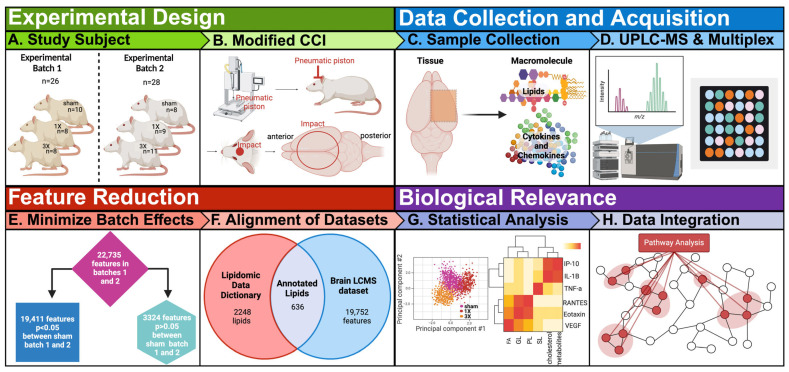
Figure 1.
Overview of study design, data acquisition, feature reduction, and biological relevance. (A) Rats were randomly assigned to different injury groups. Experimental batch 1 group were sham control group that received no injuries (n = 10), single impact group (smTBI) that received one impact (1X, n = 8), and repeat impact group (rmTBI) that received three separate impacts (3X, n = 8) with both males (n = 10) and females (n = 16). Experimental batch 2 group included sham control group that received no injuries (n = 8), smTBI group that received one impact (n = 9), and rmTBI group that received three separate impacts (n = 11) with both males (n = 17) and females (n = 11). (B) The rats received closed head impacts with a pneumatic driven controlled cortical impact (CCI), modified for closed head injury. (C) Brains were collected at 24 h post-injury and cryopulverized in preparation for lipid and cytokine analysis. (D) Samples were analyzed with Luminex multiplex assay to obtain cytokine panels and high-resolution LC-MS. Spectral alignment, peak detection and isotope and adduct grouping, gap filling, and drift correction were accomplished using Compound Discoverer v.3.0 to obtain the lipidomic dataset. (E) The features were reduced by Welch’s t-test to minimize batch effects in the lipidomic dataset. (F) The lipidomic dataset was annotated with the in-house database based on exact mass, retention time, and spectral matching. (G) Annotated lipids and cytokine panel were analyzed with volcano plots, heatmaps, charts, and principal component analysis (PCA) to understand biological significance. (H) Pathway analysis was performed to integrate the lipidomic and cytokine datasets.