Fig. 3. The structural basis of cGAS targeting by SPSB3.
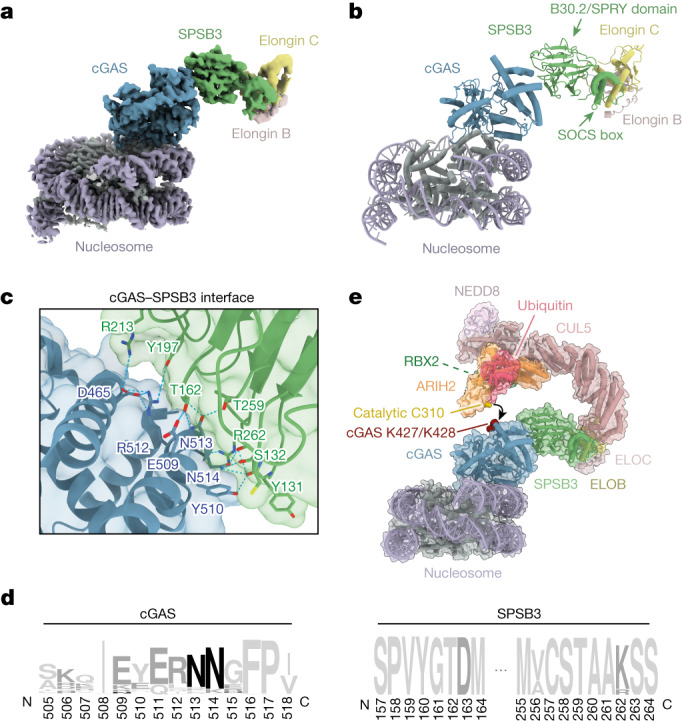
a, A composite cryo-EM density map of the nucleosome–cGAS–SPSB3–ELOBC complex, assembled from two focused-refinement maps (nucleosome–cGAS and cGAS–SPSB3–ELOBC). Different contour levels were used for optimal visualization using UCSF ChimeraX37. cGAS(site C), mutated in the DNA-binding C site, was used to obtain this dataset. b, Ribbon representation of the nucleosome–cGAS–SPSB3–ELOBC complex structure. The arrows indicate the B30.2/SPRY and SOCS box domains of SPSB3. c, Detailed view of the binding interface between cGAS and SPSB3. d, Sequence logo analysis of the cGAS and SPSB3 interface derived from 150 vertebrate species. e, Model of nucleosome-bound cGAS targeted by an activated (neddylated) CRL5–SPSB3 complex with RBR E3 ligase ARIH2 priming polyubiquitylation by transferring the first ubiquitin onto cGAS. Lysine residues that have been identified as cGAS ubiquitylation sites by MS and are essential for SPSB3-mediated degradation of cGAS are coloured red; the catalytic Cys310 of ARIH2 is coloured gold. The model was built by docking the nucleosome–cGAS–SPSB3–ELOBC model into a model comprising the ELOBC–CUL5 (Protein Data Bank (PDB): 4JGH)33, CUL5–NEDD8–RBX2–ARIH2 (PDB: 7ONI)31 and ARIH1–UB (PDB: 7B5M)32 complexes.
