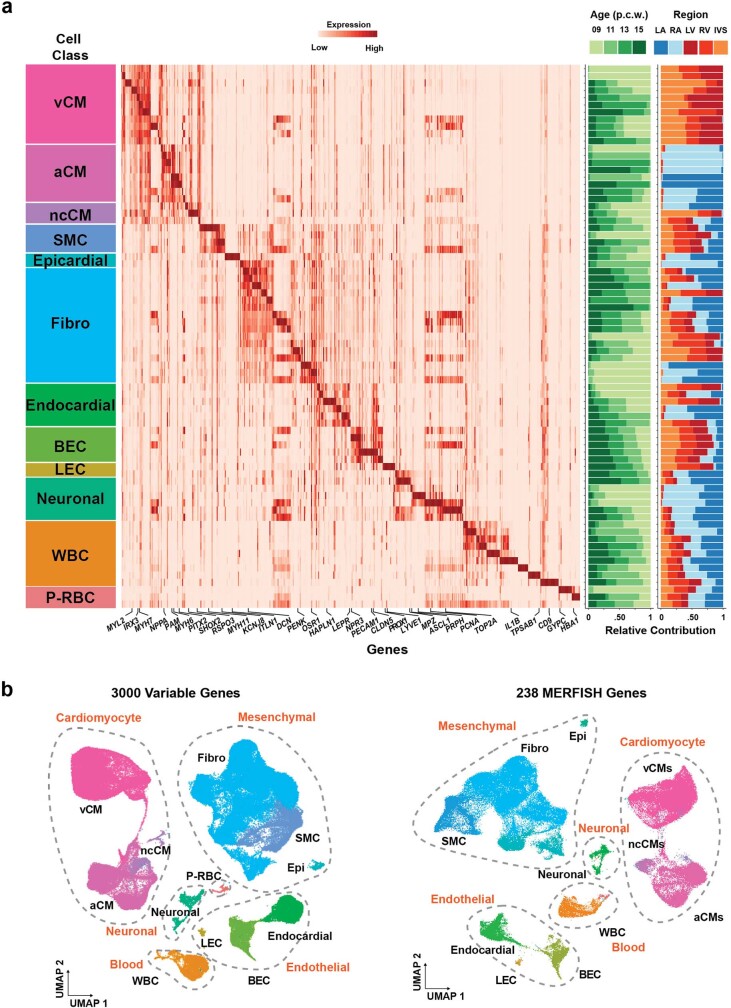
Extended Data Fig. 1. Each distinct cardiac cell identified in the scRNA-seq dataset can be molecularly defined by a limited number of genes.
a, Heatmap shows specific marker genes, as identified by NS-Forest2 classifier, for the 75 distinct cells across the developing heart. The distribution of these cells is shown according to age and region on bar graph. b, Cardiac single cells identified using the top 3,000 variable genes (left) and the 238 MERFISH genes (right) were visualized by UMAP which show that the transcriptional differences between the cell compartments (grey dashed lines) and classes (colored in a) are preserved with a limited set of genes. aCM, atrial cardiomyocyte; BEC, blood endothelial cell; Epi, epicardial; Fibro, fibroblast; IVS, interventricular septum; LA, left atrium; LEC, lymphatic endothelial cell; LV, left ventricle; ncCM, non-chambered cardiomyocyte; p.c.w., post conception weeks; P-RBC, platelet-red blood cell; RA, right atrium; RV, right ventricle; SMC, smooth muscle cell; vCM, ventricular cardiomyocyte; WBC, white blood cell.