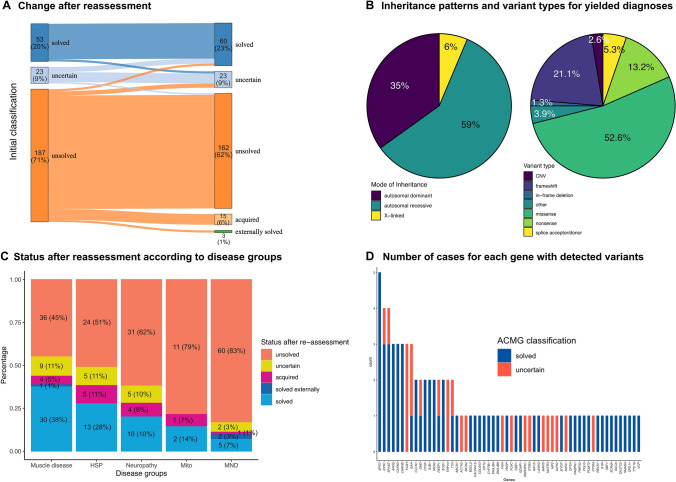
Fig. 1.
Overview of results after reassessment of NGS data. Panel A shows the change in classification of all patients after extensive reassessment including reclassification of all variants, reanalysis of all negative cases and extensive re-phenotyping and evaluation of all available clinical data including external genetic reports. Cases were regarded as solved if they had a (likely) pathogenic variant in a gene with an established gene-disease relationship compatible with the phenotype of the patient. In case of compound heterozygous variants both variants needed to be classified as (likely) pathogenic. Uncertain cases were patients where at least one of the variants in question was classified as a variant of uncertain significance (VUS). Cases were classified as having an acquired disease if NGS was negative and clinical and other diagnostic findings were compatible with a distinct non-monogenic disorder (see supplementary file 1 for a clinical description of all cases). Three patients were solved by external genetic testing after negative NGS. Panel B shows the inheritance pattern of all yielded diagnosis (excluding patients with VUS) and the type of all variants classified as (likely) pathogenic. Other variants included 1 intronic variant, 1 splice region (near-splice) variant and a GCG repeat expansion detected by WES in ABPN1. Panel C shows the classification of cases by disease (phenotype) groups after reassessment as described for panel A. Panel D shows a list of all genes with the number of detected (likely) pathogenic variants and VUS for each gene. ACMG denotes American College of Medical Genetics and Genomics, CNV copy number variation, HSP hereditary spastic paraplegia, Mito mitochondrial disease and MND motor neuron disease