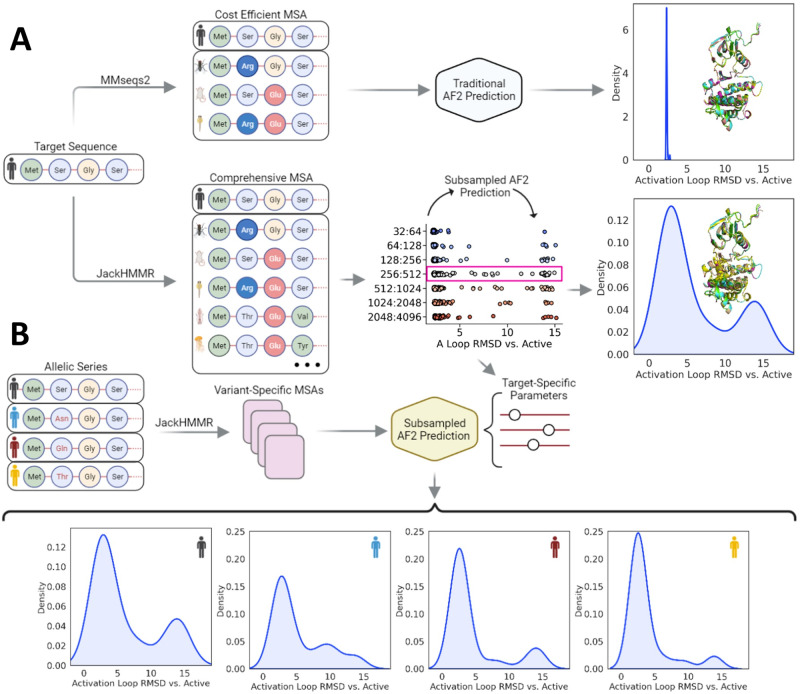
Fig. 1. Summary of the subsampled AF2 workflow employed in this study.
A Traditionally, AF2 predicts the structure of a target by using a multiple sequence alignment (MSA). When running AF2 with standard parameters, the predicted structures are often similar to each other even with a large number of independent predictions (seeds). B In this study, we show that subsampling deep MSAs causes AF2 to output predictions that occupy different conformations of the same protein, and the predicted frequency of each conformation based upon a range of random seeds strongly correlates with its experimentally-determined relative state population. Figure Created with BioRender.com.