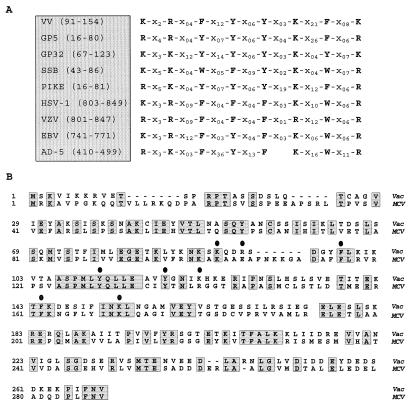
FIG. 11.
Conservation of sequence motifs within SSBs. (A) A common motif is found in diverse SSBs. Partial protein sequences from various SSBs were aligned with that portion of the gene 5 protein (fd bacteriophage) involved in DNA binding (modified from references 11, 34, and 51). This analysis yielded a region in all of the sequences which consists of conserved aromatic and basic amino acids (indicated by single-letter abbreviations) separated by variable numbers of unrelated residues. These spacer regions are designated by X, with a subscript indicating the number of residues in each case. The proteins shown are the I3 protein (39) of vaccinia virus strain WR (VV), gene 5 protein of bacteriophage fd (GP5), gene 32 protein of bacteriophage T4 (GP32), SSB of E. coli (SSB), and DNA binding proteins from bacteriophage ike (PIKE), herpes simplex virus type 1 (HSV-1), varicella-zoster virus (VZV), Epstein-Barr virus (EBV), and adenovirus type 5 (AD-5). Numbers indicate the residue numbers for each sequence shown in the alignment. (B) Sequence comparison of the vaccinia virus I3 protein and the homolog encoded by MCV. The sequence of the I3 protein encoded by the WR strain of vaccinia virus protein was aligned with the sequence of the MCV homolog (ORF MC046L, accession no. U60315). Residues common to both proteins are shaded and boxed; circles are above the residues corresponding to the motif shown in panel A.