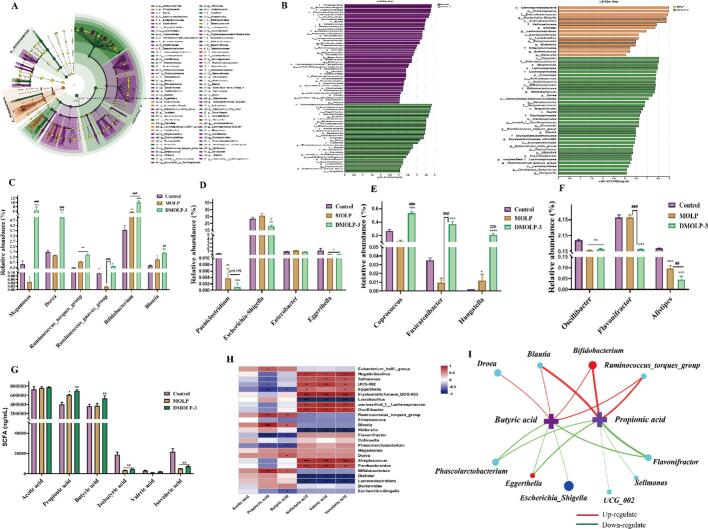
Fig. 7.
The differences in gut microbes were compared based on LEfSe and LDA scores. (A). LEfSe analysis of MOLP and DMOLP-3 at the OTUs level; (B). LDA scores among MOLP and DMOLP-3 at the OTU level; (C). Positively regulated microorganisms among MOLP and DMOLP-3 at the OTUs level; (D). Negatively regulate microorganisms among MOLP and DMOLP-3 at OTUs level; (E). Positively regulated of nerve microorganisms among the MOLP and DMOLP-3 strains at the OTU level; (F). Negatively regulate nerve-microorganisms among MOLP and DMOLP-3 at OTUs level; (G). The concentrations of SCFAs among MOLP and DMOLP-3; (H—I). Spearman Correlation analysis between genus level microorganism and SCFAs among MOLP and DMOLP-3. All the data are expressed as the means ± SEM from three independent experiments, and significance was determined using an unpaired t-test; *p < 0.05, **p < 0.01, and ***p < 0.001 vs. the control; #p < 0.05, ##p < 0.01, and ###p < 0.001 vs. the MOLP group.