FIGURE 3.
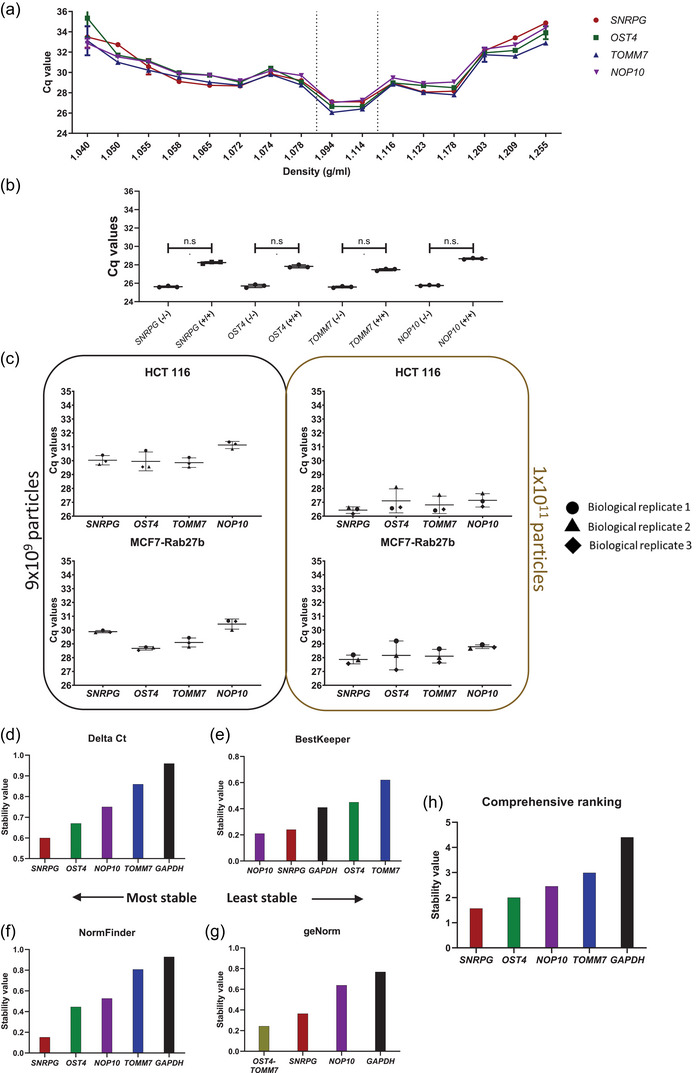
Validation of SNRPG, OST4, TOMM7 and NOP10 as suitable EV reference genes (RGs) using enzymatic treatments, biological replicates and stability algorithms. (a) RT‐qPCR analysis of all density fractions prepared from media conditioned by HCT 116 cells using DGUC. (b) RT‐qPCR analysis of SNRPG, OST4, TOMM7 and NOP10 in all density fractions prepared from media conditioned by HCT 116 cells using DGUC prior (blue) and after (red) proteinase K and RNase A treatment. Dotted lines represent average Cq values of selected RGs in soluble protein fractions (1.06–1.07 g/mL). (c) Intrinsic biological variability of EVs prepared from media conditioned by HCT 116 or MCF7 cells using DGUC in three biological replicates. Normalization to particle number was performed via nanoparticle tracking analysis (NTA). Biological results are shown as average Cq values of three technical replicates. (d–h) Stability evaluation using the RefFinder tool of EV RGs versus the established tissue and cell lines RG GAPDH in EVs prepared from media conditioned by LnCap via DGUC. Results of four algorithms are shown, (d) Delta Ct, (e) BestKeeper, (f) NormFinder and (g) geNorm alongside their respective (h) geometric mean.
