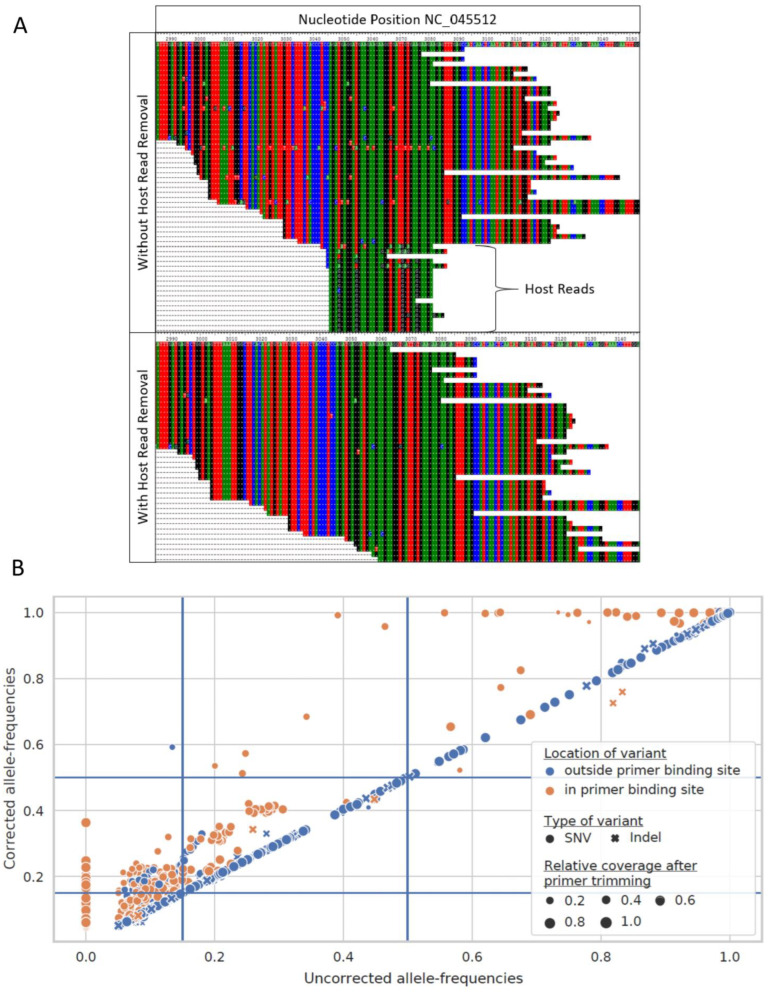
Figure 2.
The impact of host contamination removal and primer trimming. (A) The removal of host reads from RNAseq SARS-CoV-2 sequencing result, SRA run SRR12245095, reduced the potential for false-positive variant calls. In the top panel, additional mutations were present in aligned reads between positions 3049–3076 of NC_045512 when host reads were not removed. After excluding host reads (bottom panel), reads containing the mutations were no longer observed. (B) Allele frequencies of variants called after trimming primer sequences from aligned reads (corrected allele frequencies) are plotted against allele frequencies of the same variants called without primer trimming (uncorrected allele frequencies). Primer trimming increases the allele frequencies of most within-primer binding sites variants. Blue lines represent the allele-frequency thresholds used in this study to filter variant calls (allele frequency; AF ≥ 0.15) and to call consensus variants (AF ≥ 0.5).